Published online Nov 14, 2006. doi: 10.3748/wjg.v12.i42.6797
Revised: January 6, 2006
Accepted: January 14, 2006
Published online: November 14, 2006
AIM: To determine if novel bile acid transporters may be expressed in human tissues.
METHODS: SLC10A1 (NTCP) was used as a probe to search the NCBI database for homology to previously uncharacterized ESTs. The homology search identified an EST (termed SLC10A4) that shares sequence identity with SLC10A1 and SLC10A2 (ASBT). We performed Northern blot analysis and RT-PCR to determine the tissue distribution of SLC10A4. SLC10A4 was cloned in frame with an epitope tag and overexpressed in CHO cells to determine cellular localization and functional analysis of bile acid uptake.
RESULTS: Northern analysis revealed that SLC10A4 mRNA is ubiquitously expressed in human tissues with the highest levels of mRNA expression in brain, placenta, and liver. In SLC10A4-transfected CHO cells, immunoblotting analysis and immunofluorescence staining demonstrated a 49-kDa protein that is expressed at the plasma membrane and intracellular compartments. Functional analysis of SLC10A4 showed no significant taurocholate uptake in the presence of sodium when compared to untransfected CHO cells.
CONCLUSION: To date, we have shown that this protein has no capacity to transport taurocholate relative to SLC10A1; however, given its ubiquitous tissue distribution, it may play a more active role in transporting other endogenous organic anions.
- Citation: Splinter PL, Lazaridis KN, Dawson PA, LaRusso NF. Cloning and expression of SLC10A4, a putative organic anion transport protein. World J Gastroenterol 2006; 12(42): 6797-6805
- URL: https://www.wjgnet.com/1007-9327/full/v12/i42/6797.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i42.6797
Bile acids, the major solute in bile, are physiologically important for promoting bile flow and facilitating the absorption of dietary lipids[1,2]. Bile acids are also involved in cholesterol homeostasis, xenobiotic excretion, as well as apoptotic and cell signal transduction pathways[1-6]. Unlike most other biliary solutes, bile acids are efficiently conserved and cycle in the enterohepatic circulation (EHC). This EHC is maintained by the vectorial transport of bile acids via specific bile transport proteins located on the plasma membranes of liver and intestinal epithelia[1].
Within the liver, hepatocytes express the sodium dependent taurocholate transporting protein (SLC10A1) and several sodium independent organic anion-transporting proteins at the sinusoidal membrane[6-9]. On their canalicular membrane, hepatocytes express the bile salt export pump (ABCB11)[10]. Cholangiocytes, the epithelial cells that line the bile ducts, express the apical sodium dependent bile acid transporter (SLC10A2) on lumenal surface[11]. Rat cholangiocytes express a truncated form of ASBT, that may export bile acids across the basolateral membrane[12]. To complete the EHC of bile acids, the enterocytes of the distal ileum express SLC10A2[13]. After SLC10A2-mediated uptake, bile acids are directed across the ileal enterocyte and secreted into the portal circulation, allowing for efficient recycling of the bile acids from the intestine to the liver[13].
Given the redundancy of biological systems, we hypothesize that additional bile acid transporters may exist within the human genome. In this study, we identified a novel member termed SLC10A4 of the SLC10A family of sodium bile acid symporter related proteins. We also examined the expression of SLC10A4 mRNA presence in human tissues and have over expressed this protein in CHO cells.
All chemicals were of highest commercially available purity and were purchased from Sigma, St. Louis, MO Chemical C. (St. Louis, MO) unless otherwise indicated.
Cells were maintained at 37°C in a humidity-controlled incubator with CO2. The cholangiocarcinoma cell-lines (Witt, KMC, and KMCH) were grown in Dulbecco's modified Eagle’s medium containing F12 (Sigma, St. Louis, MO) supplemented with 5% (v/v) FBS (Mediatech, Herndon, VA), Penicillin (100 U/mL)/Streptomycin 100 μg/mL (Sigma, St. Louis, MO), and 2 mmol/L-glutamine (Sigma, St. Louis, MO). H69 cells are SV40 transformed normal human biliary epithelial cells (a gift from Dr. Douglas Jefferson, Tufts University) and were grown as previously described. T84 cells, derived from a human adenocarcinoma, were grown in Dulbecco’s modified Eagle’s medium containing F12 (Sigma, St. Louis, MO) supplemented with 5% (v/v) FBS (Mediatech, Herndon, VA), Penicillin (100 U/mL)/Streptomycin 100 μg/mL (Sigma, St. Louis, MO). Chinese hamster ovary (CHO) cells were grown in Dulbecco’s modified Eagle’s medium containing F12 (Sigma, St. Louis, MO) supplemented with 10% (v/v) FBS (Mediatech, Herndon, VA), Penicillin (100 U/mL)/Streptomycin 100 μg/mL (Sigma, St. Louis, MO), 1 × MEM vitamin solution (Sigma, St. Louis, MO), and 2 mmol/L L-glutamine (Sigma, St. Louis, MO).
To initially identify novel members of the SLC10 family of proteins, a bioinformatic approach was used. Amino acids 1-200 of SLC10A1 (GenBank accession no. AAA36381) were used to perform a tBlastN search of the NCBI database. The candidate EST (Htm1-361F, GenBank accession no. BE39690) sequence was then used for BlastN and BlastX database searches to confirm that it had not been previously identified and as a template to design PCR primers.
Total RNA was extracted from the tissue culture cell-lines using Tri-Reagent (Sigma, St. Louis, MO). The cells were lysed using 1 mL of Tri-Reagent/1.0 × 106 cells and stored for 5 min at room temperature. Next, 0.1 mL of 1-bromo-3-chloropropane was added, the samples were vortexed, incubated at room temperature for 15 min, and centrifuged at 12 000 × g for 15 min at 4°C. RNA was subsequently precipitated with isopropanol. The RNA pellet was resuspended with RNA Secure (Ambion, Austin, TX), and the concentration and purity was assessed by spectroscopy.
5 μg of total RNA was reverse transcribed using a SuperScriptTM II reverse transcritpase (In Vitrogen, Carlsbad, CA). The reaction mixture contained total RNA, 10 mmol/L deoxynucleotide triphosphates and random hexamers in a final volume of 10 μL. This mixture was incubated 5 min at 65°C. Reverse transcription buffer, 25 mmol/L MgCl2, 0.1 mmol/L dithiothreitol (DTT), and RNase inhibitor was then added to the reaction mixture and incubated for 2 min at 25°C. Finally, the reverse transcriptase was added to the reaction mixture and incubated for 50 min at 42°C. The reverse transcriptase reaction was terminated at 70°C for 15 min and chilled on ice. The SLC10A4 cDNA was PCR amplified using SLC10A4 specific primers; sense (5’-GGCAATCTCTCCAATCTTATGTC-3’) and antisense (5’-CAGTTGGTGGAGATGAAGAGAGT-3’). The 441 bp PCR amplicon was electrophoresed on a 1% agarose gel and the bands were visualized by ethidium bromide staining. The PCR amplicons were then cloned into the pCRII dual promoter vector (In Vitrogen, Carlsbad, CA) according to the manufacturers protocol. Briefly, 2 μL of the PCR product was mixed with 10 × ligation buffer, 50 ng of pCRII vector, water, and T4 DNA ligase. The ligation reaction was incubated overnight at 14°C and transformed into competent Escherichia coli (E. coli). Following the isolation of plasmid DNA, the pCRII (SLC10A4) was sequenced to confirm identity of the DNA insert (Mayo Molecular Core Facility, Rochester, MN).
The pCRII (SLC10A4) plasmid was linearized and random primed using radiolabeled [α-32P] dCTP. The random primer protocol was followed per manufactures (In Vitrogen, Carlsbad, CA) directions. Briefly, 25 ng of pCRII (SLC10A4) template DNA was denatured by boiling for 5 min and cooled on ice. To the template DNA, the following was added; 2 μL of dATP, 2 μL of dGTP, 2 μL of dTTP, 15 μL of random primer buffer, 5 μL of [α-32P] dCTP (10 Ci/L). This reaction mixture was brought up to 49 μL. The 1 μL of Klenow fragment was added, mixed, and centrifuged briefly. This reaction mixture was incubated for 1 h at 25°C. Following the incubation, 5 μL of stop buffer was added and the 32P-labeled SLC10A4 cDNA probe was purified using a NAP-25 column. Next, the Human MTN Blot (Clontech, Palo Alto, CA) and the Human Digestive System MTN Blot (Clontech, Palo Alto, CA) was prehybridized in ExpressHyb solution (Clontech, Palo Alto, CA) for 30 min at 68°C and then 1 × 106 cpm/mL of the labeled probe was added and incubated for an additional 60 min at 68°C. Following hybridization, the blots were washed several times and exposed to x-ray film (Kodak, Rochester, NY) and developed. Subsequently, the membranes were stripped with 0.5% SDS at 95°C for 10 min and reprobed with β-actin to normalize for mRNA loading.
We obtained IMAGE clone 3502817 (ATCC, Manassas, VA) that contains the complete ORF of SLC10A4 in the pOTB7 vector. The IMAGE clone was used as template DNA for PCR to delete the stop codon using specific PCR primers; sense (5’-GGATCCCCAAGTAACTATAACGGTCC-3’) and antisense (5’-GAATTCGTATCTCCACATTTGGAGAGAAGTCTG -3’). This fragment was cloned into pCRII (In Vitrogen, Carlsbad, CA) using the method described above. The pCRII (SLC10A4) was digested with EcoRI and BamHI restriction enzymes, the SLC10A4 fragment was isolated by gel purification, and subcloned into the EcoRI and BamHI sites of pcDNA4 V5-HisA (In Vitrogen, Carlsbad, CA), a mammalian expression vector which expresses the V5 epitope tag. The SLC10A4 insert was sequenced to confirm identity of the DNA insert (Mayo Molecular Core Facility, Rochester, MN).
Transfections were performed using Lipofectamine reagent (In Vitrogen, Carlsbad, CA) according to the manufactures directions. The day of transfection, the CHO cells were 50%-80% confluent. The media was replaced with Optimem (In Vitrogen, Carlsbad, CA) and the cells were transfected with 3 μg of pcDNA V5-HisA (SLC10A4) or empty vector pcDNA V5-HisA plasmid DNA for 4 h after which the transfection media was replaced with complete CHO media. After 48 h, Zeocin (500 μg/mL) was added to the complete media and the cells were selected. Individual stable clones were isolated using cloning cylinders and confirmed by RT-PCR and immunoblotting to confirm the expression of SLC10A4.
CHO cells were grown on collagen-coated coverslips and fixed by 0.1 mol/L PIPES, pH 6.95, 1 mmol/L ethylene glycolbis (β-amino-ethylether-N, N, N’, N’-tetra acetic acid (EGTA), and 2% paraformaldehyde in 1X phosphate buffered saline (PBS) for 20 min at room temperature. The cells were then permeabilized in 0.2% Triton for 2 min and incubated for 1 h with V5 epitope tag (1:500; In Vitrogen, Carlsbad, CA) monoclonal antibody. The cells were washed with PBS and subsequently incubated Texas Red-conjugated goat anti-mouse secondary antibody (Molecular Probes, Eugene, OR). The coverslips were washed with PBS and then mounted using Prolong Antifade mounting medium (Molecular Probes, Eugene, OR) and analyzed using a confocal microscope.
For the immunoblotting assay, cell lysates and mixed plasma membranes were prepared as previously described[14]. Cell lysates and MPM were heated to 95°C for 10 min in sample buffer containing 0.8 mol/L dithiothreitol (Sigma, St. Louis, MO) and 10% SDS (Sigma, St. Louis, MO) for protein denaturation and solubilization. These protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. After blocking, blots were incubated with the V5 epitope tag antibody (1:2000 dilution; In Vitrogen, Carlsbad, CA) and incubated over night at 4°C. The blots were washed and incubated for 1 h at room temperature with horseradish peroxidase conjugated secondary antibody (1:2000 dilution), and bands were detected using the enhanced chemiluminescent plus detection system (ECL Plus; Amersham, Arlington Heights, IL). Autoradiographs were obtained by exposing the nitrocellulose to Kodak XAR film (Rochester, NY).
Uptake experiments were performed in triplicate using un-transfected, empty vector transfected or SLC10A4-transfected CHO cells. CHO cells transfected with SLC10A1 was used as the positive control for the uptake experiments. [3H]Taurocholate (specific activity 2.0 Ci/mmol) of > 95% purity was purchased from PerkinElmer (Boston, MA). The uptake experiments were performed as previously described[15].
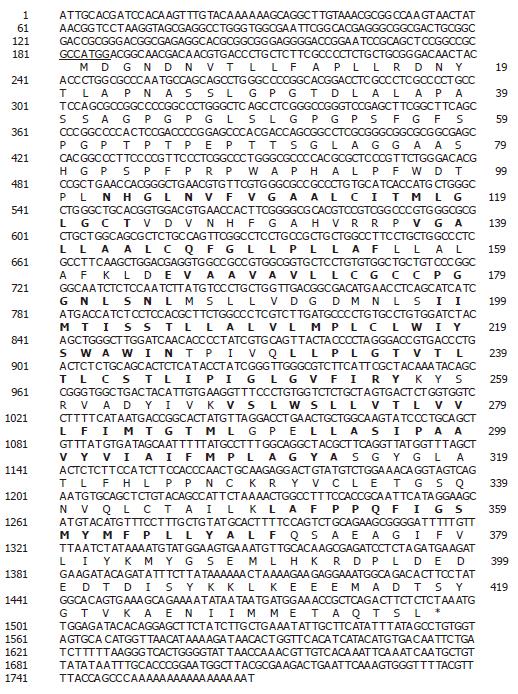
Using a bioinformatics approach, numerous expressed sequence tags (EST), including (Htm1-361f [GenBank accession no. BE439690] and Htm1-302f [GenBank accession no. BE439637]) were identified within the NCBI database. Upon further analysis it was determined that these ESTs represented a novel gene (SLC10A4) belonging to the sodium bile acid symporter family. The DNA and deduced amino acid sequence are shown in Figure 1. The human SLC10A4 cDNA encodes a protein of 437 amino acids with a predicted molecular mass of 46.5 kDa. The cDNA encompasses a consensus methionine start codon[16] and an in-frame stop codon (Figure 1). The cDNA also encompasses a 5’ untranslated region (UTR) of 183 bp and a 271 bp 3’ UTR which contains a poly A tail. Kyte-Doolittle hydropathy analysis suggests that SLC10A4 contains 8 putative transmembrane domains (Figure 1). Furthermore, the amino terminus is predicted to contain three potential N-linked glycoslylation sites.
The SLC10A4 deduced amino acid sequence for human compared to rat (accession no. XP_579196) share the greatest evolutionary conservation of SLC10A4 with 89% identity (Figure 2A). In contrast the amount of identity shared between human compared to mouse (XP_775579) or bovine (XP_591836) was equal, 73%.
To further our understanding between the relationships of SLC10A4 and the other members of the SLC10 family, we performed multiple amino acid alignments and examined the evolutionary relationship between the SLC10 proteins. At the amino acid level, SLC10A4 shares 30% identity with SLC10A3 (P3; a SLC10A family member of unknown function), 27% identity with SLC10A5, 27% identity with SLC10A2, and 24% identity with SLC10A1 (Figure 2B). We have also looked for relationships of this SLC10A4 protein compared with anion transporters but no conservation was found (data not shown).
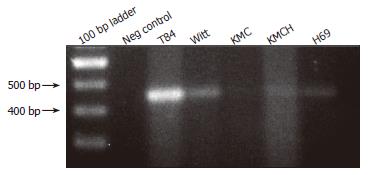
RT-PCR was performed using SLC10A4-specific primers and cDNA prepared from human colon, T84 cells, and cholangiocyte cell lines. This analysis identified SLC10A4 mRNA in T84 cells, a normal human cholangiocyte cells (H69), and the cholangiocarcinoma cell-lines Witt, KMC and KMCH (Figure 3). In each case, the PCR amplicon was sequenced to confirm the identity of the PCR product.
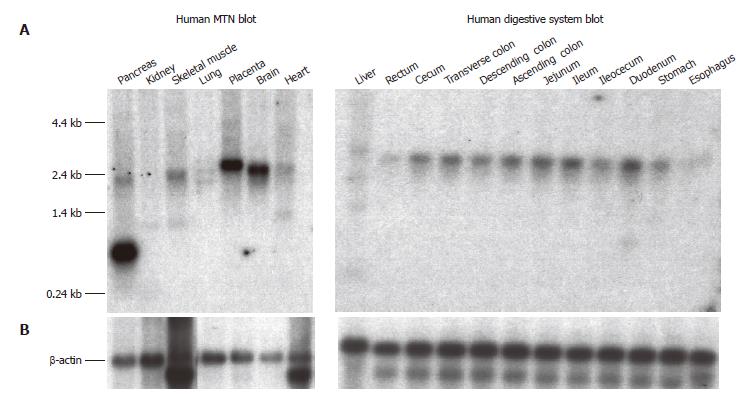
The SLC10A4 transcript was detected by Northern blot analysis in a wide variety of human tissues (Figure 4A). The highest SLC10A4 mRNA expression was observed in brain, placenta, and pancreas. Lower levels of expression were also observed in liver and kidney. While the major SLC10A4 transcript is approximately 2.4 kb, an additional prominent 0.7 kb transcript was observed in pancreas, and a minor 1.4 kb transcript was observed in liver and kidney. β-actin expression appears to be comparable in each tissue suggesting equal loading of mRNA (Figure 4B).
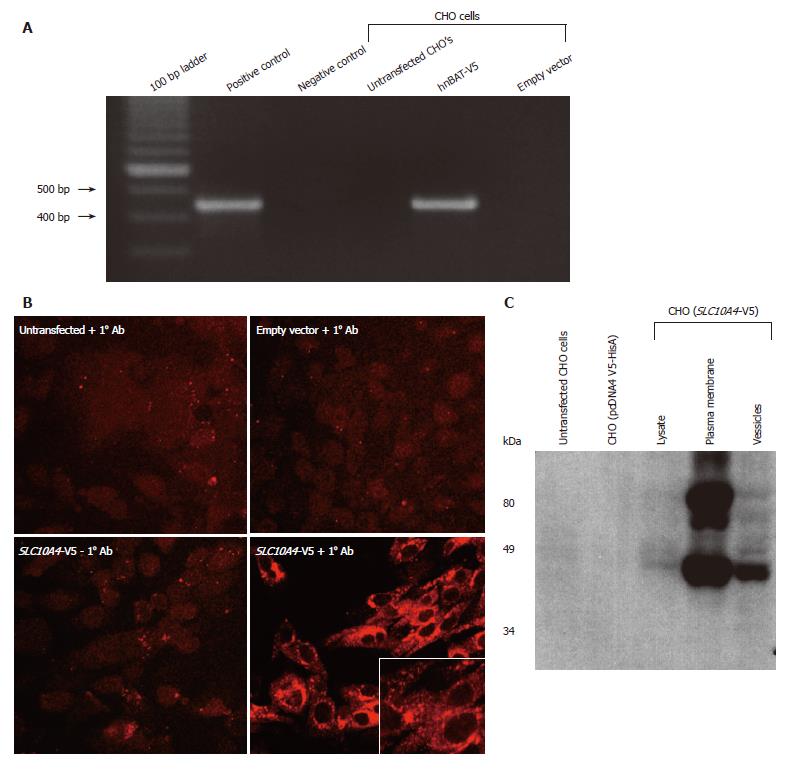
CHO cells lacking endogenous expression of SLC10A4 were stably transfected with the human SLC10A4 cDNA. Figure 5A shows that the pcDNA V5-HisA (SLC10A4) transcript is expressed in the stably transfected CHO cells. In contrast, the SLC10A4 mRNA was not detected in non-transfected CHO cells or in CHO cells transfected with the expression vector (pcDNA4 V5-HisA) alone.
To address the cellular localization of this novel protein, transfected cells were stained using an affinity-purified mouse monoclonal antibody raised against the V5 epitope tag. We observed strong staining for SLC10A4 in the transfected CHO cells, confirming its expression (Figure 5B). The transfected V5-epitope tagged SLC10A4 protein was expressed primarily in intracellular compartments and to a lesser degree on the plasma membrane.
Protein extracts from untransfected CHO cells, empty vector transfected CHO cells, and SLC10A4-transfected CHO cells were fractionated into mixed plasma membranes and an intracellular fraction and used for immunoblotting analysis. This analysis detected a 49-kDa protein and larger apparent aggregate, possibly a glycosylated form of this protein, in the SLC10A4-transfected cells, whereas no protein was detected in the non-transfected or empty vector-transfected CHO cells (Figure 5C). The 49-kDa band was detected in the lysate, plasma membrane and vesicles of the SLC10A4-transfected CHO cells.
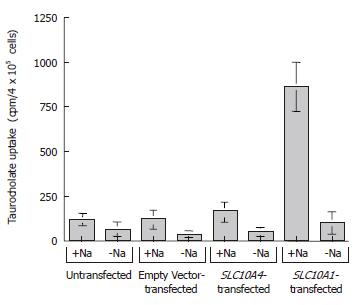
To examine the possible functional capability of SLC10A4, we performed taurocholate uptake studies using stably transfected CHO cells (Figure 6). In functional uptake the positive control, namely, CHO cells stably transfected with SLC10A1, exhibited an approximate seven fold increase in taurocholate uptake compared to untransfected and empty vector transfected cells in the presence of sodium (Figure 6). In the same experiment, SLC10A4-transfected cells showed no significant increase in taurocholate uptake when compared to the untransfected cells (Figure 6). In the absence of sodium, neither transporter (i.e. SLC10A1 or SLC10A4) showed a significant bile acid uptake when compared to control cells (Figure 6).
The major findings described here are: (a) SLC10A4 shares amino identity with other members of the sodium bile acid symporter family (SLC10A2, SLC10A1, SLC10A3, and SLC10A5); (b) the SLC10A4 mRNA is widely expressed in human tissues, including cultured human cholangiocytes; and (c) the SLC10A4 protein is expressed in intracellular compartments as well as on plasma membrane in transfected CHO cells.
As a result of the recent sequencing of the human genome, many novel genes have been identified through the cloning of ESTs (i.e., G protein coupled receptors)[17-20]. We utilized a bioinformatic approach to identify a novel SLC10A family member, termed SLC10A4. Prior to the completion of this work, two cDNA clones (accession no. AAH 12048 and AAH19066) have been sequence and listed in the NCBI database as SLC10a4 derived from neuroblastoma brain tissue. To date, only minimal bioinformatics observations have been made regarding SLC10A4[21]; however to our knowledge this is the first report to have experimentally analyzed SLC10A4 mRNA distribution in various tissues, including cholangiocytes. Furthermore we have expressed the SLC10A4 protein in a CHO cell-line to determine its subcellular localization.
The SLC10A4 amino acid sequence is conserved amongst various mammalian species. This suggests that this protein may have an important role in substrate transport within various mammal species. SLC10A4 protein has a long extracellular amino terminus (shared amongst species) relative to that of SLC10A1 and SLC10A2 which may suggest a unique physiological process and role that this protein may have.
SLC10A4 shares many characteristics of the human sodium dependent bile acid symporter family members. The SLC10 family is composed of five family members that show approximately 30% identity at the amino acid level. Within the SLC10 family, SLC10A1 and SLC10A2 share the greatest amino acid identity (about 36%) and are the only proteins shown to transport bile acids. From an evolutionary perspective, SLC10A4 appears to be an intermediate between SLC10A1 and the uncharacterized SLC10A5 protein. To this end, the exon structure of the sodium dependent bile acid symporter family is of interest. SLC10A5 has 1 predicted exon; SLC10A4 and SLC10A3 are composed of 3 predicted exons, SLC10A1 is encoded by 5 exons, and SLC10A2 is encoded by 6 exons. The simpler genomic organization of SLC10A4, SLC10A5 and SLC10A3 allows us to hypothesize that the genes may be ancesteral in nature. SCL10A4 is composed of 437 amino acids compared to SLC10A1 and SLC10A2 that are made of 349 and 348 amino acids, respectively. Furthermore, SLC10A1 and SLC10A2 are glycosylated and SLC10A4 is predicted to have numerous glycosylation sites on the amino terminus. The predicted size of SLC10A4 is slightly smaller than the experimental molecular weight, which may be due to addition of the V5 epitope tag on its carboxyl terminus. Additionally, the immunoblot shows a larger band of about 80 kDa that is possibly due to the glycosylation of the native protein, which is expected to occur based on the bioinformatics predictions. In contrast to SLC10A1 and SLC10A2, the hydropathy plot for SLC10A4 suggests eight potential transmembrane-spanning domains whereas the more characterized models of SLC10A1 and SLC10A2 have only seven again suggesting that this protein may have diverged from the well studied SLC10A1 and SLC10A2 genes. This discrepancy will require further experimental analysis to determine the actual number of transmembrane domains that reside in SLC10A4. The major sodium bile acid symporters involved in the secretion and absorption of bile acids are SLC10A1 and SLC10A2. SLC10A1 is primarily found in the liver where it is localized in the hepatocytes[6]. Within the hepatocytes, the SLC10A1 protein resides on the sinusoidal membrane[6]. In contrast, SLC10A2 mRNA has been shown in the cholangiocytes, ileal enterocytes, and in the proximal tubular cells of the kidneys[11,15,22,23]. SLC10A2 is an apical oriented protein in the afore mentioned cells. Both SLC10A1 and SLC10A2 have been shown to transport various bile acids in a sodium dependent manner[6,15,22,23]. Much less is known about the remaining members of the SLC10 family of sodium bile acid symporters. SLC10A3 is ubiquitously expressed in tissues and has been localized to various ESTs including placenta[21] (GenBank accession no. BX377672, brain (GenBank accession no. BM559383), lung (GenBank accession No. BM981524), kidney (GenBank accession no. BG249893) and stomach (GenBank accession no. BM747336). Far less is known for SLC10A5 mRNA tissue distribution that has been limited to fetal brain (GenBank accession no. XM_294493)[21]. SLC10A4 is also ubiquitously expressed in all tissues we have tested. Interestingly, SLC10A4 mRNA shows slight difference in mRNA size amongst tissue by Northern blotting suggesting that this may be the result of tissue specific alternative transcription.
To our knowledge, the protein localization and substrate specificity have not been determined for either SLC10A3 or SLC10A5, and these genes remain orphan transport proteins. In CHO cells expressing SLC10A4, we were not able to show transport of taurocholate, suggesting that SLC10A4 is also orphan transport protein.
We have found that the SLC10A4 mRNA is widely expressed in human tissues and is comparable to distribution pattern of SLC10A3. In fact, both SLC10A3 and SLC10A4 mRNA are expressed in similar tissues, suggesting both proteins may serve a ubiquitous function in the cell machinery. To define the localization of SLC10A4 in CHO cells we created a chimeric protein in which the coding sequence of SLC10A4 was cloned in frame with the V5-epitope tag. This construct was stably transfected in CHO cells to characterize the SLC10A4 protein. We found that the novel protein localizes to both intracellular and plasma membrane of CHO cells. The intracellular localization infers that the protein is a regulated protein by some intracellular messenger. It has been shown the SLC10A1-GFP is regulated by cAMP when this construct was transfected into HepG2 cells[24]. It is possible that SLC10A4 is regulated in a similar manner as SLC10A1. SLC10A4 may be a regulated protein that requires an agonist to translocate the protein to the plasma membrane.
The physiological relevance of this novel transporter is currently unclear. Although the protein does not transport taurocholate and chenodeoxycholic acid, we cannot rule the transport capacity of other bile acids or other polar solutes. Nevertheless, SLC10A4 protein shares homology with SLC10A1 and SLC10A2. We have cloned and characterized the human SLC10A4 in hoped that they data will improve our understanding on the phylogeny, origin and evolution of the SLC10A family of transport proteins which may provide important insight into which protein regions are involved in substrate specificity.
We thank XM Chen and PS Tietz for helpful discussions and D Hintz for secretarial assistance.
S- Editor Wang J L- Editor Alpini GD E- Editor Liu WF
| 1. | Hofmann AF. Intestinal absorption of bile acids and biliary constituents: the intestinal component of the enterohepatic circulation and the integrated system. Physiology of the gastrointestinal tract. Raven: New York 1994; 1845-1865. |
| 2. | Baxter JD, Webb P. Metabolism: bile acids heat things up. Nature. 2006;439:402-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001;29:1467-1472. [PubMed] |
| 4. | Jones BA, Rao YP, Stravitz RT, Gores GJ. Bile salt-induced apoptosis of hepatocytes involves activation of protein kinase C. Am J Physiol. 1997;272:G1109-G1115. [PubMed] |
| 5. | Mühlbauer M, Allard B, Bosserhoff AK, Kiessling S, Herfarth H, Rogler G, Schölmerich J, Jobin C, Hellerbrand C. Differential effects of deoxycholic acid and taurodeoxycholic acid on NF-kappa B signal transduction and IL-8 gene expression in colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:G1000-G1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest. 1994;93:1326-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 338] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 7. | Abe T, Kakyo M, Tokui T, Nakagomi R, Nishio T, Nakai D, Nomura H, Unno M, Suzuki M, Naitoh T. Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J Biol Chem. 1999;274:17159-17163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 373] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 8. | Kullak-Ublick GA, Hagenbuch B, Stieger B, Schteingart CD, Hofmann AF, Wolkoff AW, Meier PJ. Molecular and functional characterization of an organic anion transporting polypeptide cloned from human liver. Gastroenterology. 1995;109:1274-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 287] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 9. | König J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol Gastrointest Liver Physiol. 2000;278:G156-G164. [PubMed] |
| 10. | Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 674] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 11. | Que FG, Phan VA, Phan VH, LaRusso NF, Gores GJ. GUDC inhibits cytochrome c release from human cholangiocyte mitochondria. J Surg Res. 1999;83:100-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Lazaridis KN, Tietz P, Wu T, Kip S, Dawson PA, LaRusso NF. Alternative splicing of the rat sodium/bile acid transporter changes its cellular localization and transport properties. Proc Natl Acad Sci USA. 2000;97:11092-11097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 83] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Wong MH, Oelkers P, Craddock AL, Dawson PA. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J Biol Chem. 1994;269:1340-1347. [PubMed] |
| 14. | Tietz P, Levine S, Holman R, Fretham C, LaRusso NF. Characterization of apical and basolateral plasma membrane domains derived from cultured rat cholangiocytes. Anal Biochem. 1997;254:192-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Craddock AL, Love MW, Daniel RW, Kirby LC, Walters HC, Wong MH, Dawson PA. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am J Physiol. 1998;274:G157-G169. [PubMed] |
| 16. | Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2301] [Cited by in RCA: 2822] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 17. | Lee DK, Nguyen T, Lynch KR, Cheng R, Vanti WB, Arkhitko O, Lewis T, Evans JF, George SR, O'Dowd BF. Discovery and mapping of ten novel G protein-coupled receptor genes. Gene. 2001;275:83-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Wittenberger T, Schaller HC, Hellebrand S. An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. J Mol Biol. 2001;307:799-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 152] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Parker RM, Copeland NG, Eyre HJ, Liu M, Gilbert DJ, Crawford J, Couzens M, Sutherland GR, Jenkins NA, Herzog H. Molecular cloning and characterisation of GPR74 a novel G-protein coupled receptor closest related to the Y-receptor family. Brain Res Mol Brain Res. 2000;77:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Lee DK, Lynch KR, Nguyen T, Im DS, Cheng R, Saldivia VR, Liu Y, Liu IS, Heng HH, Seeman P. Cloning and characterization of additional members of the G protein-coupled receptor family. Biochim Biophys Acta. 2000;1490:311-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Hagenbuch B, Dawson P. The sodium bile salt cotransport family SLC10. Pflugers Arch. 2004;447:566-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 189] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Lazaridis KN, Pham L, Tietz P, Marinelli RA, deGroen PC, Levine S, Dawson PA, LaRusso NF. Rat cholangiocytes absorb bile acids at their apical domain via the ileal sodium-dependent bile acid transporter. J Clin Invest. 1997;100:2714-2721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 184] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 23. | Kip NS, Lazaridis KN, Masyuk AI, Splinter PL, Huebert RC, LaRusso NF. Differential expression of cholangiocyte and ileal bile acid transporters following bile acid supplementation and depletion. World J Gastroenterol. 2004;10:1440-1446. [PubMed] |
| 24. | Dranoff JA, McClure M, Burgstahler AD, Denson LA, Crawford AR, Crawford JM, Karpen SJ, Nathanson MH. Short-term regulation of bile acid uptake by microfilament-dependent translocation of rat ntcp to the plasma membrane. Hepatology. 1999;30:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 55] [Article Influence: 2.1] [Reference Citation Analysis (0)] |