INTRODUCTION
Nuclear factor κB (NF-κB) is a family of pleiotropic transcription factors[1]. It regulates the transcription of a large number of genes that play key roles in embryonic development, lymphoid differentiation, apoptosis, and immune and inflammatory responses[2-5] .They are characterized by the presence of so called Rel homology domain, RHD, with a length of about 300 amino acids. Their active DNA-binding forms are homodimeric or heterodimeric complexes consisting of combinations of these protein family members. The most abundant forms of NF-κB are p65/p50 hetero-dimers and p65/p65 homo-dimers[6-7]. In most cells, NF-κB complexes are normally localized to the cytosol as inactive complexes with inhibitory IκBα protein[8]. Activation of NF-κB in response to stimuli involves activation of IκB kinase (IKK), phosphorylation and degradation of IκBα at two serine residues (Ser32 and Ser36), followed by rapid ubiquitin-dependent degradation by the 26S proteosome and release of activated NF-κB[9-11]. Activated NF-κB then translocates to the nucleus, where it binds to its target DNA sequence and activates the transcription of a vast number and wide range of genes[12-15].
RelA, the p65 subunite of NF-κB is constitutively activated in certain neoplastic cells, such as pancreatic cancer cells and acute leukemia cells[16-19]. Approaches to suppress NF-κB activation in malignant cells have been considered as a potential treatment for neoplasia. Studies show that inhibition of NF-κB activation by expression of a dominant-negative mutant form IκBα (Ser 32, 36A) (IκBαM) completely inhibited liver metastasis of a pancreatic cancer cell line, and reduced angiogenesis in an ovarian cancer cell line[20-22].
Recent studies have shown that IκBα is found to inhibit p53 tumor suppressor protein by binding p53 to form a cytoplasmic p53· IκBα complex, thus it prevents p53 nuclear translocation[23]. On the basis of this data, we hypothesized that when IκBα or its mutants were used to mediate activities of NF-κB in cells, they might affect the p53 signaling pathway simultaneously. In this study, pECFP-IκBα and its three mutants, pECFP-IκBαM (amino acides 1-317, S32, 36A), pECFP-IκBα243N (N terminus of IκBα, amino acides 1-243) and pECFP-IκBα244C (C terminus of IκBα, amino acides 244-317), were constructed. The location patterns of NF-κB, p53 and IκBα (IκBαM, IκBα243N, IκBα244C) were observed by laser confocal scanning microscopy. The effects of IκBα (IκBαM, IκBα243N, IκBα244C) on p53 and NF-κB, as well as the downstream genes of these two signaling pathways, were studied with real time QT-PCR. The relationship of NF-κB, p53, and IκBα activities is further discussed.
MATERIALS AND METHODS
Materials
Mammalian cell expression vector pEYFP-p65 was provided by Professor Johannes A. Schmid[24]. A human full-length IκBα cDNA was found in the universal GenBank database (gene number: AY033600) and was obtained from Funeng company (vector: RB01-IκBα). pDsRed-Mit vector was provided by Dr. Fuminori Tsuruta[25]. Wild-type p53 cDNA was provided by Dr. Ye KH (Jinan University, Guangzhou). Dulbecco’s modified Eagle medium (DMEM) was purchased from GIBCO (Grand Island, NY). The RNA isolation kit and LightCycler FastStart DNA Master SYBR GreenIkit were obtained from Roche. M-MLV Reverse Transcriptase was provided by BBI. LipofectamineTM Reagent was purchased from Invitrogen. Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan).
Construction of CFP-IκBα, YFP-p65 and p53-DsRed variants
Four expression constructs were constructed with the pECFP-C1 vector (cloning site, EcoRI and BamHI; Clontech): (1) IκBα, the entire coding region (amino acides 1-317)(primers: FW3/RV1); (2) IκBαM, dominant negative IκBα construct made by altering Ser-32 to Ala-32 and Ser-36 to Ala-36, (amino acides 1-317, Ser32A,Ser36A) using primers FW1/RV1, FW2/RV1 and FW3/RV1 in turn; (3) IκBα243N, the N-terminal ankyrin region (amino acides 1-243 ) (primers: FW3/RV2); (4) IκBα244C, the C-terminal domain (amino acids 244-317) (primers: FW4/RV1). Wild-type p53 cDNA were cloned into the NheI and BamHI sides of pDsRed-Mit vector (primers: p53F/p53R). The synthetic primers used for making these constructs by PCR are FW1: 5’-gag cgg cta ctg gac gac cgc cac gac gcc ggc ctg gac gcc atg aaa gac gag gag ta-3’, FW2: 5’-g gag tgg gcc atg gag ggc ccc cgc gac ggg ctg aag aag gag cgg cta ctg gac gac c-3’, FW3: 5’-c cgg aat tca ttc cag gcg gcc gag cgc ccc cag gag tgg gcc atg gag ggc c-3’, FW4: 5’-gg aat tct aac aga gtt acc tac cag ggc ta-3’, RV1: 5’-cgc gga tcc tca taa cgt cag acg ctg gcc tcc aaa cac aca gtc -3’,RV2: 5’-cg gga tcc tta tca atg gtg atg gtg atg gtg gac atc agc ccc aca ctt-3’, p53F: 5’-c tag cta gcg gaa gct tcc acc atg gag gag ccg cag tca gat-3’, p53R: 5’- c ggg atc ccg gtc tga gtc agg ccc ttc tgt-3’. All constructs were verified by restriction and sequence analysis.
Cell culture and transfections
Cell line, ASTC-α-1, was cultured in DMEM medium, supplemented with HEPES and 100 mL/L new born calf serum, and maintained at 37°C at an atmosphere of 5% CO2. Transient transfections were performed using the LipofectamineTM Reagent (Invitrogen). Cells were transfected with CFP-C1 as a control. Microscopy of cells, RNA extraction and RT were performed 30 h after transfection.
Laser scanning microscopy
YFP-p65, p53-DsRed and CFP-IκBα were visualized by using a laser scanning microscope (LSM510/ConfoCor2, Zeiss, Jena, Germany) with a 37°C stage incubator. The distribution of YFP-p65 was observed by 514nm laser (HFT458/514, LP530). Cells transfected with p53-DsRed were observed with a 543nm laser and fluorescent images were collected with a 560 nm long-pass filter (HFT700/543, NFT545, LP560nm). CFP-IκBα, CFP-IκBαM, CFP-IκBα243N and CFP-IκBα243C were observed with an Argon-ion laser with 458 nm output and a band pass barrier filter (HFT458 nm, NFT545 nm, BP470-500 nm).
RNA extraction and RT
Total RNA was isolated by using a high purity RNA isolation kit (Roche) according to the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed with 20 U of M-MLV Reverse Transcriptase, using Oligo(dT)18 primers (BBI) according to the manufacturer’s instructions.
LightCycler real-time QT-PCR
cDNA amplification by QT-PCR was carried out with the LightCycler FastStart DNA Master SYBR GreenI kit (Roche). For QT-PCR, a mastermix of the following reaction components was prepared: 0.8 μL MgCl2 stock solution, (25 mmol/L), 2 μL LightCycler FastStart DNA Master SYBR Green I, 2 μL (0.3 µmol/L) each of the primers, 11.2 μL water. LightCycler mastermix was filled in the glass capillaries and 2 μL of total cDNA temple was added. PCR primers were target gene 1 (p65: forward primer, 5’- GGCTATAACTCGCCTA GTGA -3’; reverse primer, 5’- CGAAG GAGCTGATCTGACTCA -3’), gene 2 (NF-κB downstream gene, TNF-α[26]: forward primer, 5’- CAGAGG GAAGAGTTCCCCAG -3’; reverse primer, 5’- CCTTGGTCTGGTAGGA GACG -3’), gene 3 (p53: forward primer, 5’-AGGTTGGCTCTGACTGTA-3’; reverse primer, 5’- GCAGCTCGTGGTGAGGCTC -3’), and gene 4 (p53 downstream gene, Bax[27]: forward primer, 5’- CTGACA TGTTTTC TGACGGC -3’; reverse primer, 5’-TCAGCCCATCTTCTTCCAGA-3’). In all experiments, β-actin was the reference (forward primer, 5’-GAAATCGTGCGTGACATTAA-3’; reverse primer, 5’- GGA CTCGTCATACTCCTG-3’).
The following LightCycler experimental run protocol was used: denaturation program (95°C for 10 min), amplification and quantification program repeated 40 times (95°C for 10 s, 55°C for 5 s, 72°C for 10 s), melting curve program (65-95°C with a heating rate of 0.1°C per second and a continuous fluorescence measurement) and finally a cooling step to 40°C. For the mathematical model it is necessary to determine the crossing points (CP) for each transcript. CP is defined as the point at which the fluorescence rises appreciably above the background fluorescence. The ‘Fit Point Method’ must be performed, at which CP will be measured at a constant fluorescence level[28]. Results are expressed as the target/reference ratio of the sample divided by the target/reference ratio of the control.
CCK-8 experiment
Different transfected group cells were cultured in 96-well microplates for 48 h. CCK-8 was added to the cells and incubated for 2 h. OD450, the absorbance value at 450 nm, was read with a microplate reader (DG5032, Hua dong, Nanjing, China). The value is directly proportional to the number of viable cells in a culture medium and the cell proliferation.
Statistical analysis
Statistical results were obtained using the statistical software SPSS. The significant difference tests were based on analysis of variance with a single factor and two sample t-tests were performed.
RESULTS
Localization patterns of p53-DsRed, YFP-p65, CFP-IκBα and its mutants in living cells
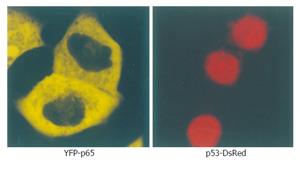
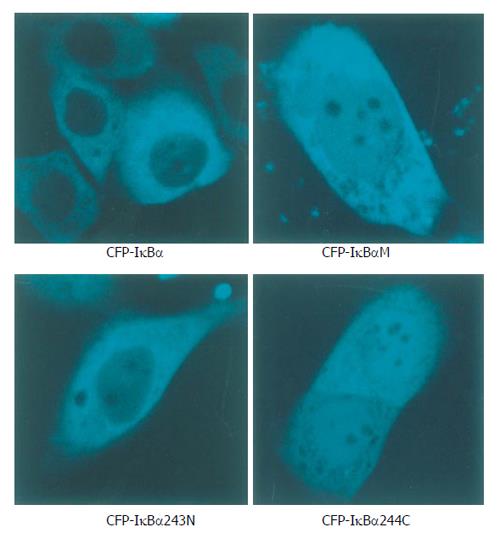
Cells transfected with p53-DsRed revealed a predominant nuclear localization. YFP-p65 mainly existed in the cytoplasm (Figure 1). Cells were transfected with CFP-IκBα, CFP-IκBαM and CFP-IκBα243N respectively, and revealed a predominant cytosolic localization, while cells transfected with CFP-IκBα244C revealed a predominant nuclear localization of CFP-IκBα244C (Figure 2).
Figure 1 Microscopy of YFP-p65 and p53-DsRed in living cells.
Figure 2 Localization patterns of CFP-IκBα, CFP-IκBαM, CFP-IκBα243N and CFP-IκBα244C in living cells.
Standard curve for real time QT-PCR
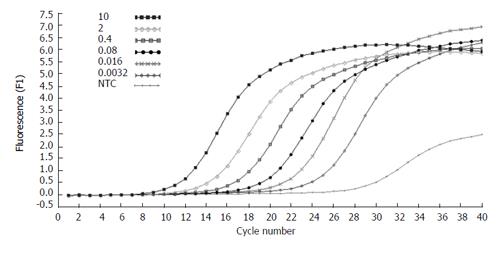
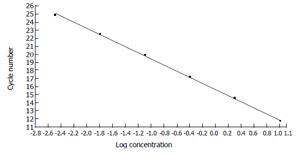
The concentration of the standards covers the expected concentration range of all samples. Dilution folds of the cDNA template for the standard curve run were 10ul to 3.20E-3 μL (Figures 3 and 4). The standard curves were analyzed with Real Quant Software to create a coefficient file. The coefficient file was used later in the relative quantification analysis.
Figure 3 Amplification plots of five fold serial dilutions of β-actin cDNA.
The fluorescence values versus cycle number are displayed.
Figure 4 Standard curve constructed with the β-actin cDNA standards from 1.
00E + 1 to 3.20E + 3 by plotting the logarithmic concentration of the standard versus the crossing points (cycle number).
Effects of IκBα and its mutants on the NF-κB signaling pathway
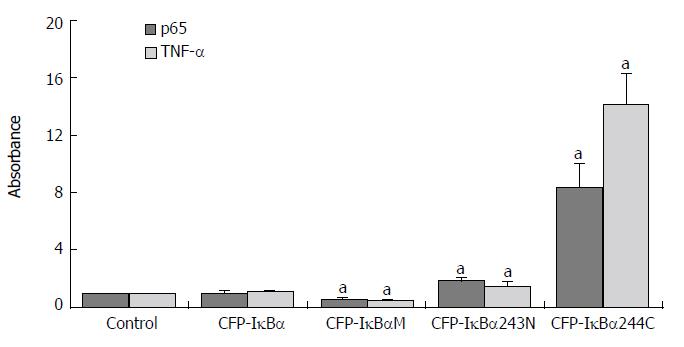
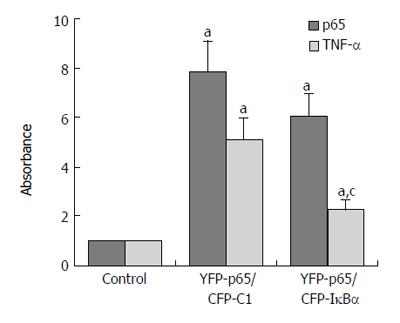
Results are expressed as the target/reference ratio of the samples divided by the target/reference ratio of the control (n = 3). In all experiments β-actin cDNA was the reference. Results for the analysis of different transfected cells by QT-PCR showed that the level of p65 cDNA/β-actin cDNA (0.945 ± 0.152) and TNF-α cDNA /β-actin cDNA (1.05 ± 0.106) in CFP-IκBα transfected cells did not change significantly compared with the control(1.000 ± 0.000) (Figure 5), while in YFP-p65/CFP-IκBα co-transfected cells, IκBα decreased the transcription of p65 downstream gene TNF-α (2.24 ± 0.503) compared with the YFP-p65/CFP-C1 co-transfected cells(5.08 ± 0.891) (cP < 0.05) (Figure 6). In CFP-IκBαM transfected cells, the transcription level of the two genes (0.548 ± 0.086, 0.53 ± 0.056) decreased compared with the control (aP < 0.05). The level of p65 cDNA /β-actin cDNA and TNF-α cDNA /β-actin cDNA in CFP-IκBα243N transfected cells increased a bit (1.84 ± 0.176, 1.51 ± 0.203) (aP < 0.05). The most prominent was CFP-IκBα244C. It increased the transcription level of all the genes significantly (8.29 ± 1.662, 14.16 ± 2.121) compared with the control (aP < 0.05) (Figure 5).
Figure 5 Effects of IκBα and its mutants on NF-κB and NF-κB downstream gene TNF-α.
Abscissa showed different transfected Cells. Y-coordinate expressed the target/reference ratio of the samples divided by the tar = get/reference ratio of the control. In all experiments β-actin cDNA was reference. (aP < 0.05 vs control).
Figure 6 Effects of IκBα on over expressed NF-κB.
(aP < 0.05 vs control; cP < 0.05 vs YFP-p65/CFP-C1 group).
Effects of IκBα and its mutants on the p53 signaling pathway
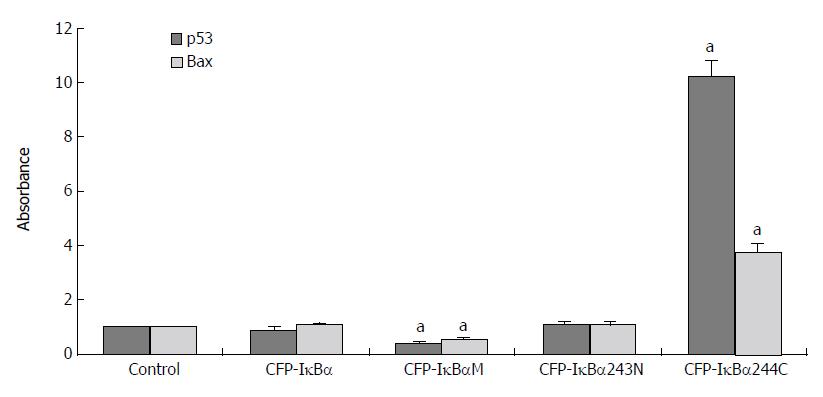
Results for the analysis of different transfected cells by QT-PCR indicated that the effect of IκBα and its mutants on p53 and its downstream gene, Bax, were different (Figure 7). The level of p53 cDNA /β-actin cDNA and Bax cDNA /β-actin cDNA in CFP-IκBα (0.9 ± 0.126, 1.04 ± 0.109) and CFP-IκBα243N (0.806 ± 0.129, 0.79 ± 0.108) transfected cells did not change very much. In CFP-IκBαM transfected cells, the transcription level of the genes decreased (0.43 ± 0.061, 0.53 ± 0.063) compared with the control (aP < 0.05), however, CFP-IκBα244C increased the transcription level of p53 and Bax significantly (10.2 ± 0.621, 3.72 ± 0.346) (aP < 0.05) (Figure 7), which suggested that IκBα244C may play an important role in inducing apoptosis[14].
Figure 7 Effects of IκBα and its mutants on p53 and p53 downstream gene Bax.
(aP < 0.05 vs control).
IκBα244C and p53 synergistically mediates apoptosis
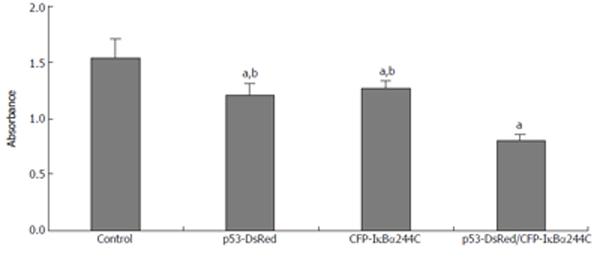
To study the effect of IκBα244C on cell death, a CCK-8 experiment was performed. As Figure 8 shows, transient expression of p53-DsRed (1.206 ± 0.099 ) or CFP-IκBα244C (1.259 ± 0.072) resulted in enhancement of cell death compared with the control (1.531 ± 0.168) (n = 6, aP < 0.05). The synergistic effect in mediating apoptosis by p53-DsRed/CFP-IκBα244C (0.805 ± 0.047) (dP < 0.01) was obtained.
Figure 8 IκBα244C and p53 synergistically mediates apoptosis.
(aP < 0.05 vs control; bP < 0.01 vs p53-DsRed/CFP-IκBα244C group).
DISCUSSION
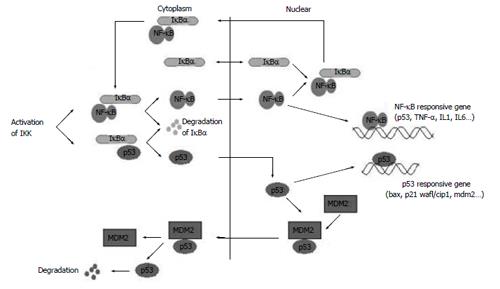
NF-κB and p53 are important transcription factors present in the majority of cells[27-30]. Constitutively activated NF-κB has been associated with increased cell proliferation and survival in cancer cells. Inhibitor of NF-κB alpha, IκBα, participates in both NF-κB and p53 signaling pathways[23,31-33] (Figure 9). The functional NF-κB and p53 activities may modulate each other, which in turn could affect the subsequent responses.
Figure 9 IκBα participates in NF-κB and p53 signaling pathways.
Previous studies demonstrated that IκBα interacts with NF-κB and p53 with different interaction sides[2,8,23]. IκBα and its mutants might have different effects on the transcription of NF-κB, p53 and their downstream genes, according to their different structures. Our studies showed that IκBα did not influence the transcription level of NF-κB, p53 and their downstream target genes in static cells compared with controls, which maybe due to the integrity of IκBα and the self-regulation capability of the cells. IκBα243N (amino acides 1-243), with lack of the PEST domain that regulates basal level protein turnover and is required for inhibition of DNA binding of NF-κB, increased the transcription of NF-κB and TNF-α slightly. Because IκBα243N cannot interact with p53, it has no effect on the transcription of p53 and Bax.
Dominant negative IκBα (IκBαM, Ser32, 36A) and the C terminus of IκBα (IκBα244C, amino acides 244-317) are notable because of their significantly different effects. IκBαM has mutations in Ser32 and Ser36. It can not phosphorylate at Ser32 and Ser36 and degradate, so IκBαM bound with NF-κB and p53 in the cytoplasm steadily and inhibited the transcription of their downstream genes, which is consistent with the report that IκBαM has been found to represses p53-dependent apoptosis in acute lymphoblastic leukemia cells[34]. In particular, transfection of IκBαM in human colon carcinoma and breast cancer cell lines did not increase sensitivity to daunomycin or Taxol[35,36]. IκBαM may repress p53 expression in two ways: (1) A portion of IκBαM directly interacts with p53 in cytoplasm and inhibits p53 translocate to the nucleus; (2) IκBαM binds to NF-κB in the cytoplasm and NF-κB·IκBαM complex is formed, which in turn inhibits the NF-κB activity and the NF-κB dependent p53 activity, for the NF-κB signaling cascade is a potential modulator of p53 activity, and NF-κB is a co-factor of p53 in mediating cell death[37-39].
IκBα244C does not have the ARD (ankyrin repeat domain) and NES in N terminus. It could not prevent NF-κB from translocating to the nucleus, and IκBα244C itself mainly existed in the nucleus. IκBα244C enhanced the transcription level of p53, NF-κB and their downstream genes. The CCK-8 experiment showed that co-expression of p53 with IκBα244C resulted in enhancement of p53-mediated cell death. p53 and IκBα244C are possibly co-factors in inducing apoptosis, and the C terminus of IκBα may serve as a pro-apoptotic protein in living cells.
NF-κB has been considered a target for cancer treatment[17,40]. The function of IκBα as an inhibitor in regulating NF-κB activation has been well studied. Findings from the present study suggest that mutants of IκBα have different effects on NF-κB and p53 signaling pathways, and may result in different therapy results. The inhibition effect of IκBαM indicates drugs that induce apoptosis by a p53-dependent mechanism may be inhibited by the use of IκBαM constructs through inhibition of p53 function by these agents. The C terminal of IκBα enhanced cell death, which suggests that it may be a pro-apoptotic protein existing in cells, but the mechanism remains to be determined and there may exist NF-κB and p53 independent pathways.