Published online Oct 28, 2006. doi: 10.3748/wjg.v12.i40.6540
Revised: July 12, 2006
Accepted: September 13, 2006
Published online: October 28, 2006
AIM: To determine the genotypes in Mexican hepatitis B virus (HBV) isolates and characterize their precore and core promoter mutations.
METHODS: Forty-nine HBV isolates of Mexico obtained from sera of 15 hepatitis patients, 6 hemodialysis patients, 20 men seeking HIV testing, and 8 AIDS patients were analyzed. HBV isolates were amplified by PCR, and genotyped by line probe assay (INNO-LiPA HBV Genotyping; INNOGENETICS N V, Ghent, Belgium). HBV genotype confirmation was performed by DNA sequencing part of the sAg region. Precore and core promoter mutation characterization was performed by line probe assay (INNO-LiPA HBV PreCore; INNOGENETICS N V, Ghent, Belgium).
RESULTS: Overall, HBV genotype H was found in 37 (75.5%) out of the 49 isolates studied. HBV genotypes G, A, and D were found in 5 (10.2%), 4 (8.2%), and 3 (6.1%) isolates, respectively. HBV genotype H was predominant in isolates from hemodialysis patients (100%), hepatitis patients (80%), and men seeking HIV testing (75%), and accounted for half of infections in AIDS patients (50%). Six (12.2%) out of the 49 HBV isolates showed both wild type and mutant populations at precore codon 28. These mixed wild type and precore mutant populations were observed in one HBV genotype A isolate and in all HBV genotype G isolates. A dual variant core promoter mutation was observed in 1 (2%) of the isolates, which was genotype H.
CONCLUSION: HBV genotype H is highly predominant in HBV isolates of Mexico followed by genotypes G, A and D. A low frequency of precore and core promoter mutations is observed in HBV Mexican isolates.
- Citation: Alvarado-Esquivel C, Sablon E, Conde-González CJ, Juárez-Figueroa L, Ruiz-Maya L, Aguilar-Benavides S. Molecular analysis of hepatitis B virus isolates in Mexico: Predominant circulation of hepatitis B virus genotype H. World J Gastroenterol 2006; 12(40): 6540-6545
- URL: https://www.wjgnet.com/1007-9327/full/v12/i40/6540.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i40.6540
Hepatitis B virus (HBV) is an important cause of morbidity and mortality worldwide. It is estimated that 2 billion people are infected with HBV and 350 million individuals suffer from chronic HBV infection in the world[1,2]. Chronic HBV infection may lead to hepatic fibrosis, cirrhosis and hepatocellular carcinoma (HCC)[3]. In addition, HBV infection is the 10th leading cause of death worldwide[1]. HBV is a hepadnavirus that possesses a double stranded DNA genome[4]. The genetic organization of HBV has been described elsewhere[4]. Briefly, HBV genome consists of four partly overlapping open reading frames: S that encodes for envelope proteins (HBsAg), C for core protein (HBcAg) and e antigen (HBeAg), P for polymerase protein and X for transcriptional transactivator protein[4]. Based on sequence divergence of the entire genome of > 8%, HBV genomes have been classified into 8 genotypes designated A to H[5,6]. The distribution of HBV genotypes is geographically restricted. In the American continent, HBV genotypes A and C are common in the USA[7,8], while genotype F is predominant in Central American countries[9]. In Europe, HBV genotype D is prevalent in Mediterranean Europe[10], while genotype A is frequent in Northwest Europe[11]. In the African continent, HBV genotype E is common in the Sub-Saharan Africa[12], and genotype A in South Africa[13]. In the Asian continent, HBV genotypes B and C predominate in south east countries[14-16], while genotype D is prevalent in central Asia[17]. Reports indicate that HBV genotypes are related with the severity of liver disease. HBV genotype C has been associated with the development of liver cirrhosis and hepatocellular carcinoma[14,18-21]. In addition, HBV genotypes are related with the response to antiviral therapy. Asian studies have shown that compared to HBV genotype B, genotype C has a lower response to interferon alpha (IFN-α) therapy[22,23]. A recent European randomized trial showed that patients infected with HBV genotypes A and B respond significantly better to pegylated IFN-α-2b alone or in combination with lamivudine than patients infected with HBV genotypes C and D[24]. Mutations in the precore and core promoter regions of HBV have been also related with genotypes. Precore and core promoter mutations have been observed mainly in HBV genotypes D and B[25-27], as well as A and C[25,28,29], respectively.
Little is known about the molecular epidemiology of HBV in Mexico. A previous study on HBV genotypes published in 1998 showed that HBV genotype F is predominant in Mexico[30]. However, more recently it was discovered that HBV genotype F is divergent[31], to the extent that a new genotype is split off from genotype F, this new genotype has been designated as HBV genotype H[9]. Thus some divergent strains formerly classified as HBV genotype F could now be classified as HBV genotype H. Therefore, in order to obtain an updated classification we sought to determine the HBV genotypes and characterize the precore and core promoter mutations in HBV isolates of Mexico.
Forty-nine HBV isolates of Mexico were analyzed. HBV isolates were obtained from serum samples of 15 hepatitis patients, 6 hemodialysis patients, 20 men seeking HIV antibody testing, and 8 AIDS patients. All samples were collected in Mexico City and Guadalajara City; both cities are located in central Mexico. Participants were of Mestizo ethnicity. All samples were HBsAg positive. HBeAg, anti-HBe, and HBV DNA levels were not determined. Men seeking HIV antibody testing came from a high risk population for sexually transmitted infection acquisition. Seropositivity for anti-HBc among them (30.4%) was associated to men who had sex with men exclusively and seropositivity for both HIV and herpes simplex virus type 2[32].
HBV DNA was extracted from 200 μL of serum by using the high pure PCR template preparation kit (Roche Applied Science, Penzberg, Germany) as recommended by the manufacturer. HBV DNA was amplified with HBV pol gene domain B and C primers (INNO-LiPA HBV DR amplification, INNOGENETICS N. V., Ghent, Belgium) by nested PCR as described in the product insert. Primer sequences were used as previously described[33]. PCR conditions were as follows: 40 cycles at 94°C for 30 s, at 45°C for 30 s and at 72°C for 30 s. HBV genotyping was performed by INNO-LiPA HBV genotyping (INNOGENETICS N. V., Ghent, Belgium) following the manufacturer’s instructions.
Sequence analysis was done directly on the second round PCR products using the big dye terminator V3.1 cycle sequencing kit of Applied Biosystems. Sequencing PCR was performed for 25 cycles (at 95°C for 10 s, at 50°C for 5 s, at 60°C for 4 min) and purified on a sephadex column. Three μL dextran blue/deionised formamide loading buffer was added to the dried pellet and 1 μL was then loaded on a 4.5% acrylamide slab gel. After electrophoresis of 14 h on the automated sequencer ABI377, data analysis was done using software sequencher 4.1.2.
HBV DNA was additionally amplified with basal core promoter and precore primers (INNO-LiPA HBV PreCore amplification, INNOGENETICS N. V., Ghent, Belgium) by nested PCR as recommended by the manufacturer. Primer sequences were used as previously described [25]. PCR conditions were as follows: 40 cycles at 94°C for 30 s, at 50°C for 30 s and at 72°C for 30 s. Characterization of precore mutations was performed by INNO-LiPA HBV precore (INNOGENETICS N. V., Ghent, Belgium) according to the manufacturer’s instructions. This method could identify nucleotide polymorphism at nt 1762 and nt 1764 in the basal core promoter and at codon 28 in the precore region of HBV[34].
For all the 49 HBV isolates, INNO-LiPA HBV genotyping assay was able to provide a genotype result. HBV genotype H was found in 36 (73.5%) out of the 49 isolates studied, while HBV genotypes G, A, D and F were found in 5 (10.2%), 4 (8.2%), 3 (6.1%) and 1 (2.0%) isolates, respectively.
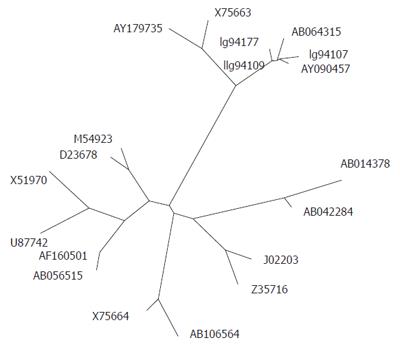
Thirty-two out of the 36 isolates of HBV genotype H, all 5 isolates of genotype G, and only 1 isolate of genotype F genotyped by INNO-LiPA were selected for DNA sequencing confirmation. All 32 isolates of genotype H and all 5 isolates of genotype G were confirmed by phylogenetic analysis of the HBsAg sequences. However, sequencing and subsequent phylogenetic analysis of the HBV genotype F showed a genotype H. This was caused by a single nucleotide mismatch with the probe on line 11 of the INNO-LiPA HBV genotyping, abolishing the reaction with this probe. This in turn led to a single reactivity with the genotype F probe on line 15. When we compared the HBV genotype H sequences obtained in this study with other published sequences by means of the phylogenetic program PHYLIP, we found that our sequences formed two separate branches within genotype H (Figure 1).
Thus, the final adjusted prevalences of HBV genotypes were 37 isolates (75.5%) of genotype H, 5 isolates (10.2%) of genotype G, 4 isolates (8.2%) of genotype A, and 3 isolates (6.1%) of genotype D. HBV genotype H was predominant in the hepatitis patients, men seeking HIV testing, and hemodialysis patients, and accounted for half of infections in AIDS patients (Table 1). Since the number of HBV isolates analyzed in some groups was very small, we were unable to provide statistical evidence for the distribution of HBV genotypes within the groups.
| Group of subjects | n | Genotypes, n (%) | |||
| A | D | G | H | ||
| Hepatitis patients | 15 | 1 (6.7) | 2 (13.3) | 0 | 12 (80) |
| Men seeking HIV testing | 20 | 3 (15) | 1 (5) | 1 (5) | 15 (75) |
| AIDS Patients | 8 | 0 | 0 | 4 (50) | 4 (50) |
| Hemodialysis patients | 6 | 0 | 0 | 0 | 6 (100) |
| Total | 49 | 4 (8.2) | 3 (6.1) | 5 (10.2) | 37 (75.5) |
In the precore region, 41 (83.7%) isolates showed wild type (G) sequence at nucleotide 1896, 6 (12.2%) had a mixture of wild type and G1896A mutation, and 2 had an indeterminate sequence (Table 2). Mixed wild type and precore mutant populations were observed in only one isolate of HBV genotype A but in all isolates of HBV genotype G. Statistical analysis showed that the frequency of mixed wild type and precore mutant populations was significantly higher (P = 0.04) in isolates of HBV genotype G than in isolates of HBV genotype A. Mixed wild type and precore mutant populations were observed in 10% of isolates from men seeking HIV testing and in 50% of isolates from AIDS patients only. The frequency of these mixed wild type and precore mutant populations in AIDS patients was significantly higher than that in isolates from hepatitis patients (P = 0.007), and men seeking HIV testing (P = 0.03). This frequency of mixed wild type and precore mutant populations in AIDS patients was also higher but not statistically significant (P = 0.06) than that in isolates from hemodialysis patients.
| Region and variant | n (%) | Genotypes | ||||
| A | D | G | H | |||
| (n = 4) | (n = 3) | (n = 5) | (n = 37) | |||
| Precore (nt 1896) | ||||||
| Wild type (G) | 41 | (83.7) | 3 | 2 | 36 | |
| Wild type + variant (G + A) | 6 | (12.2) | 1 | 5 | ||
| Indeterminate | 2 | (4.1) | 1 | 1 | ||
| Core promoter (nt 1762 and 1764) | ||||||
| Wild type (AG) | 44 | (89.8) | 4 | 2 | 5 | 33 |
| Dual variant (TA) | 1 | (2) | 1 | |||
| Indeterminate | 4 | (8.2) | 1 | 3 | ||
In the core promoter region, 44 (89.8%) isolates had wild type sequence (A at nucleotide 1762 and G at nucleotide 1764), 1 (2%) showed the classical dual variant (A1762T, G1764A), and 4 (8.2%) had an indeterminate sequence. The only one dual variant was found in a sample of a hepatitis patient infected with HBV genotype H. For its part, all 4 samples with indeterminate sequences were found in hepatitis patients too. Three of them were found in isolates of HBV genotype H, and in one isolate of HBV genotype D.
In this work, we found a predominant frequency of HBV genotype H in Mexican isolates. The extremely high frequency of this HBV genotype H in Mexico has not been reported before. A previous study in Mexican isolates from blood donors and patients with liver disease showed infections with only 3 HBV genotypes: A, D and F in both populations[30]. Nevertheless, in the present study we have found 4 HBV genotypes. It was known that HBV genotype F was predominant in Mexico [30], but also it was observed that the predominant genotype F in Mexico is divergent[31]. These results contrast with those found in the present study since HBV genotype H was predominant whilst HBV genotype F was not present anymore. Genotype H which was discovered recently is closely related with genotype F, while genotype H is most likely split off from genotype F[9]. Therefore, a number of HBV isolates previously classified as genotype F are now classified as genotype H. Thus, it explains the remarkable change in HBV genotype distribution in Mexico with a predominant circulation of genotype H. The high frequency of HBV genotype H in this study is the highest reported worldwide. Indeed, this genotype has been rarely found or not reported at all in most countries[35-39]. HBV genotype H has been found only in HBV isolates of Nicaragua, Mexico, USA[9,31,35], and Japan[37,39]. In our study, HBV genotype H was predominant in the hepatitis patients, men seeking HIV testing, and hemodialysis patients. In addition, genotype H was responsible of 50% of infections in AIDS patients. Reports on genotypes found in hepatitis patients from other countries remark the differences in geographical distribution of HBV genotypes in the world. In the USA and other countries a high frequency of HBV genotype B and C has been found in hepatitis patients[40,41]. HBV genotypes B and C have been linked to Asian ethnicity[7,35], and the absence of these genotypes in our populations studied reflects the absence of HBV isolates of Asian origin. Frequencies of HBV genotypes found in men seeking HIV antibody testing are difficult to compare since there are not further reports for the comparison. Concerning hemodialysis patients, HBV genotype H was responsible for all infections in our study. This prevalence is clearly different from those found in hemodialysis patients of other countries. For instance, in a Turkish study all isolates of hemodialysis patients were genotype D[42], in an Indonesian study all isolates were genotype B[43], and in a Brazilian study genotypes A and D were the most frequently found[44]. These HBV genotypes reported in hemodialysis patients certainly reflect the predominant HBV genotypes found in their respective countries. For its part, HBV genotype G is responsible for half of infections in AIDS patients. HBV genotype G has been discovered in USA and France isolates by Stuyver et al[45]. Interestingly, all HBV genotype G isolates were found as single isolates (no co-infections) in our study. This finding is unexpected, since HBV genotype G is frequently found as a co-infection with HBV genotype A[46,47]. The HBV genotype frequencies in AIDS patients found in our study differ substantially from those found in a Spanish study where researchers found a predominant HBV genotype A followed by genotype D among the AIDS patients studied[48]. The low frequency of precore mutants in Mexican isolates found in the present study confirms a previous observation[30]. In addition, this frequency of precore mutations contrasts with a higher frequency observed in the USA and Hong Kong[7,34,49]. The distribution of precore mutations in HBV genotypes found in the present study differs substantially from those found in other studies. While other researchers have found precore mutations in HBV genotypes B, C[50,51], and D[27], we have found the mutations only in HBV genotypes G and A, being mutations largely predominant in HBV genotype G (all HBV genotype G isolates in this study). The existence of precore mutations in HBV genotype G has been reported in patients chronically infected with HBV[45]. Remarkably, all mutated HBV genotype G isolates in this study came from samples of AIDS patients and one of the men seeking HIV testing. No similar findings are reported in AIDS patients. Concerning mutations in the core promoter region, it is remarkable that the classical dual variant (A1762T, G1764A), and indeterminate sequences were observed only in hepatitis patients. The prevalence of core promoter mutations and its distribution in HBV genotypes found in this study also differ from those reported in other countries. A large study with Hong Kong and USA samples showed that the frequency of dual variant is 20% and 30.2%, respectively[34], which is significantly higher than that found in our study. HBV core promoter mutations have been found in HBV genotypes A, B C and D[25,29,34]. Nevertheless, these mutations were found mainly in HBV genotype H followed by HBV genotype D in the present study. Indeterminate sequences in precore and core promoter regions as well as the change in genotype F in LiPA to genotype H found in the Mexican isolates are most probably due to mismatching of the sequences with the probes used. A single mismatch is sufficient to abolish hybridization to a probe under the conditions of the assay as previously described[34].
The INNO-LiPA HBV genotyping and precore assays have proved to be rapid, sensitive and reliable for the detection of HBV genotypes and precore promotor/precore mutations.
The genetic variability of HBV has implications on the sensitivity of immunologic and molecular based assays[5]. In addition, HBV genotyping has shown its utility not only in epidemiology studies but also in predicting prognosis and therapeutic response[6,52,53].
In conclusion, HBV circulates in at least 4 different genotypes in Mexico. HBV genotype H is highly predominant in HBV isolates of Mexico followed by genotypes G, A and D. The frequency of precore mutations in Mexican HBV isolates is low and associated mainly with HBV genotype G. Core promoter mutations seem to be rare in HBV isolates of Mexico.
S- Editor Wang J L- Editor Wang XL E- Editor Ma WH
| 1. | Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1734] [Cited by in RCA: 1750] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 2. | Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV. Global epidemiology of hepatitis B virus. J Clin Gastroenterol. 2004;38:S158-S168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 403] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 3. | Thomson EC, Main J. Advances in hepatitis B and C. Curr Opin Infect Dis. 2004;17:449-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Zuckerman JN, Zuckerman AJ. Infectious Diseases. Barcelona, Spain: Mosby 2000; 4.1-4.12. |
| 5. | Weber B. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J Clin Virol. 2005;32:102-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 6. | Kramvis A, Kew M, François G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409-2423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 262] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 7. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS Jr, Luketic VA, Terrault N. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 229] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 8. | Moriya T, Kuramoto IK, Yoshizawa H, Holland PV. Distribution of hepatitis B virus genotypes among American blood donors determined with a PreS2 epitope enzyme-linked immunosorbent assay kit. J Clin Microbiol. 2002;40:877-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. [PubMed] |
| 10. | Echevarria JM, Avellon A, Magnius LO. Molecular epidemiology of hepatitis B virus in Spain: identification of viral genotypes and prediction of antigenic subtypes by limited sequencing. J Med Virol. 2005;76:176-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Bielawski KP, Dybikowska A, Lisowska-Charmuszko U, Stalke P, Gregorowicz K, Trocha H, Podhajska A. Distribution of HBV genotypes and mutants among hepatitis B infected patients from northern Poland. Int J Mol Med. 2004;14:301-304. [PubMed] |
| 12. | Mulders MN, Venard V, Njayou M, Edorh AP, Bola Oyefolu AO, Kehinde MO, Muyembe Tamfum JJ, Nebie YK, Maiga I, Ammerlaan W. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J Infect Dis. 2004;190:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Kimbi GC, Kramvis A, Kew MC. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J Gen Virol. 2004;85:1211-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 14. | Zhu B, Luo K, Hu Z. [Establishment of a method for classification of HBV genome and it's application]. Zhonghua Shiyan He Linchuang Bingduxue Zazhi. 1999;13:309-313. [PubMed] |
| 15. | Liu CJ, Kao JH, Chen PJ, Lai MY, Chen DS. Molecular epidemiology of hepatitis B viral serotypes and genotypes in taiwan. J Biomed Sci. 2002;9:166-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Ishikawa K, Koyama T, Masuda T. Prevalence of HBV genotypes in asymptomatic carrier residents and their clinical characteristics during long-term follow-up: the relevance to changes in the HBeAg/anti-HBe system. Hepatol Res. 2002;24:1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 17. | Kato H, Ruzibakiev R, Yuldasheva N, Hegay T, Kurbanov F, Achundjanov B, Tuichiev L, Usuda S, Ueda R, Mizokami M. Hepatitis B virus genotypes in Uzbekistan and validity of two different systems for genotyping. J Med Virol. 2002;67:477-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Chan HL, Tsang SW, Wong ML, Tse CH, Leung NW, Chan FK, Sung JJ. Genotype B hepatitis B virus is associated with severe icteric flare-up of chronic hepatitis B virus infection in Hong Kong. Am J Gastroenterol. 2002;97:2629-2633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Ding X, Park YN, Taltavull TC, Thung SN, Jin X, Jin Y, Trung NS, Edamoto Y, Sata T, Abe K. Geographic characterization of hepatitis virus infections, genotyping of hepatitis B virus, and p53 mutation in hepatocellular carcinoma analyzed by in situ detection of viral genomes from carcinoma tissues: comparison among six different countries. Jpn J Infect Dis. 2003;56:12-18. [PubMed] |
| 20. | Lee CM, Chen CH, Lu SN, Tung HD, Chou WJ, Wang JH, Chen TM, Hung CH, Huang CC, Chen WJ. Prevalence and clinical implications of hepatitis B virus genotypes in southern Taiwan. Scand J Gastroenterol. 2003;38:95-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 387] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 22. | Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol. 2000;33:998-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 350] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 23. | Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425-1430. [PubMed] |
| 24. | Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 904] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 25. | Grandjacques C, Pradat P, Stuyver L, Chevallier M, Chevallier P, Pichoud C, Maisonnas M, Trepo C, Zoulim F. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J Hepatol. 2000;33:430-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 167] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 26. | Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepat. 2002;9:52-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 265] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 27. | Kondili LA, Brunetto MR, Maina AM, Argentini C, Chionne P, La Sorsa V, Resuli B, Mele A, Rapicetta M. Clinical and molecular characterization of chronic hepatitis B in Albania: a country that is still highly endemic for HBV infection. J Med Virol. 2005;75:20-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Chan HL, Hussain M, Lok AS. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology. 1999;29:976-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 199] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 29. | Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 315] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 30. | Alvarado-Esquivel C, Wyseur A, Herrera-Ortiz FM, Ruiz-Maya L, Ruiz-Astorga R, Zarate-Aguilar A, Carrillo-Maravilla E, Herrera-Luna R, Morales-Macedo MC, Maertens G and Stuyver L. Therapies for Viral Hepatitis. London: International Medical Press 1998; 35-41. |
| 31. | Sánchez LV, Maldonado M, Bastidas-Ramírez BE, Norder H, Panduro A. Genotypes and S-gene variability of Mexican hepatitis B virus strains. J Med Virol. 2002;68:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Juarez-Figueroa LA, Uribe-Salas FJ, Conde-Glez CJ, Hernandez-Avila M, Hernandez-Nevarez P, Uribe-Zuniga P, del Rio-Chiriboga C. Hepatitis B markers in men seeking human immunodeficiency virus antibody testing in Mexico City. Sex Transm Dis. 1997;24:211-217. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Stuyver L, Van Geyt C, De Gendt S, Van Reybroeck G, Zoulim F, Leroux-Roels G, Rossau R. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000;38:702-707. [PubMed] |
| 34. | Hussain M, Chu CJ, Sablon E, Lok AS. Rapid and sensitive assays for determination of hepatitis B virus (HBV) genotypes and detection of HBV precore and core promoter variants. J Clin Microbiol. 2003;41:3699-3705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 35. | Kato H, Gish RG, Bzowej N, Newsom M, Sugauchi F, Tanaka Y, Kato T, Orito E, Usuda S, Ueda R. Eight genotypes (A-H) of hepatitis B virus infecting patients from San Francisco and their demographic, clinical, and virological characteristics. J Med Virol. 2004;73:516-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 649] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 37. | Ohnuma H, Yoshikawa A, Mizoguchi H, Okamoto H. Characterization of genotype H hepatitis B virus strain identified for the first time from a Japanese blood donor by nucleic acid amplification test. J Gen Virol. 2005;86:595-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 38. | Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Mahboudi F, Sabahi F, Adeli A, Noorinayer B, Azizi M, Reza Zali M. Genotype characterization and phylogenetic analysis of hepatitis B virus isolates from Iranian patients. J Med Virol. 2005;75:227-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Shibayama T, Masuda G, Ajisawa A, Hiruma K, Tsuda F, Nishizawa T, Takahashi M, Okamoto H. Characterization of seven genotypes (A to E, G and H) of hepatitis B virus recovered from Japanese patients infected with human immunodeficiency virus type 1. J Med Virol. 2005;76:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 40. | Wai CT, Fontana RJ, Polson J, Hussain M, Shakil AO, Han SH, Davern TJ, Lee WM, Lok AS. Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 166] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 42. | Bozdayi AM, Aslan N, Bozdayi G, Türkyilmaz AR, Sengezer T, Wend U, Erkan O, Aydemir F, Zakirhodjaev S, Orucov S. Molecular epidemiology of hepatitis B, C and D viruses in Turkish patients. Arch Virol. 2004;149:2115-2129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Lusida MI, Surayah , Sakugawa H, Nagano-Fujii M, Soetjipto , Mulyanto , Handajani R, Boediwarsono , Setiawan PB, Nidom CA. Genotype and subtype analyses of hepatitis B virus (HBV) and possible co-infection of HBV and hepatitis C virus (HCV) or hepatitis D virus (HDV) in blood donors, patients with chronic liver disease and patients on hemodialysis in Surabaya, Indonesia. Microbiol Immunol. 2003;47:969-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | Teles SA, Martins RM, Vanderborght B, Stuyver L, Gaspar AM, Yoshida CF. Hepatitis B virus: genotypes and subtypes in Brazilian hemodialysis patients. Artif Organs. 1999;23:1074-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67-74. [PubMed] |
| 46. | Kato H, Orito E, Gish RG, Sugauchi F, Suzuki S, Ueda R, Miyakawa Y, Mizokami M. Characteristics of hepatitis B virus isolates of genotype G and their phylogenetic differences from the other six genotypes (A through F). J Virol. 2002;76:6131-6137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 102] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Kato H, Orito E, Gish RG, Bzowej N, Newsom M, Sugauchi F, Suzuki S, Ueda R, Miyakawa Y, Mizokami M. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology. 2002;35:922-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 48. | Pérez-Olmeda M, Núñez M, García-Samaniego J, Ríos P, González-Lahoz J, Soriano V. Distribution of hepatitis B virus genotypes in HIV-infected patients with chronic hepatitis B: therapeutic implications. AIDS Res Hum Retroviruses. 2003;19:657-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 49. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS Jr, Luketic VA, Terrault N. Prevalence of HBV precore/core promoter variants in the United States. Hepatology. 2003;38:619-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 156] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 50. | Sakurai M, Sugauchi F, Tsai N, Suzuki S, Hasegawa I, Fujiwara K, Orito E, Ueda R, Mizokami M. Genotype and phylogenetic characterization of hepatitis B virus among multi-ethnic cohort in Hawaii. World J Gastroenterol. 2004;10:2218-2222. [PubMed] |
| 51. | Huy TT, Ushijima H, Quang VX, Ngoc TT, Hayashi S, Sata T, Abe K. Characteristics of core promoter and precore stop codon mutants of hepatitis B virus in Vietnam. J Med Virol. 2004;74:228-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 52. | Locarnini SA. Clinical relevance of viral dynamics and genotypes in hepatitis B virus. J Gastroenterol Hepatol. 2002;17 Suppl 3:S322-S328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Wang Y, Zhou G, Li X, Zhou Z, Zhou S, Ruan L, Chen M, Deng W. [Genotyping of hepatitis B virus and clinical investigation]. Zhonghua Shiyan He Linchuang Bingduxue Zazhi. 2002;16:367-369. [PubMed] |