Published online Oct 7, 2006. doi: 10.3748/wjg.v12.i37.6032
Revised: June 22, 2006
Accepted: July 7, 2006
Published online: October 7, 2006
AIM: To evaluate the potential association of xeroderma pigmentosum group D (XPD) codon 751 variant with outcome after chemo-radiotherapy in patients with resected gastric cancer.
METHODS: We used PCR-RFLP to evaluate the genetic XPD Lys751Gln polymorphisms in 44 patients with stage III (48%) and IV (20%) gastric cancer treated with surgery following radiation therapy plus 5-fluorouracil/leucovorin based chemotherapy.
RESULTS: Statistical analysis showed that 75% (12 of 16) of relapse patients showed Lys/Lys genotype more frequently (P = 0.042). The Lys polymorphism was an independent predictor of high-risk relapse-free survival from Cox analysis (HR: 3.07, 95% CI: 1.07-8.78, P = 0.036) and Kaplan-Meir test (P = 0.027, log-rank test).
CONCLUSION: XPD Lys751Gln polymorphism may be an important marker in the prediction of clinical outcome to chemo-radiotherapy in resected gastric cancer patients.
- Citation: R RZ, Arias F, Bandres E, Cubedo E, Malumbres R, García-Foncillas J. Xeroderma pigmentosum group D 751 polymorphism as a predictive factor in resected gastric cancer treated withchemo-radiotherapy. World J Gastroenterol 2006; 12(37): 6032-6036
- URL: https://www.wjgnet.com/1007-9327/full/v12/i37/6032.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i37.6032
The xeroderma pigmentosum group D (XPD) gene encodes a protein required for nucleotide excision repair (NER). This product recognizes and repairs a wide range of structurally unrelated lesions such as bulky adducts caused by UV light, environmental agents, cross links and oxidative damage[1,2]. Moreover, XPD gene is one of the components of basal transcription factor IIH (TFIIH), participating also in transcription initiation. Since XPD is involved in both transcription and NER, it may be able to repair other types of damage, such as radiation therapy-induced damage. Because XPD interacts with many different proteins as part of TFIIH transcription factor, amino acid variants in different domains of XPD, such as 683 and 751, may affect different protein interactions, and result in expression of different phenotypes[3,4].
Intrinsic and acquired resistance to cancer chemotherapeutic agents results from polymorphisms in genes encoding DNA repair enzymes. Differences in responsiveness of cancer cells to anticancer agents can be differently affected by changes in repair efficiency. Clinical outcomes after chemotherapy may be influenced by pharmacogenetic polymorphisms in DNA repair enzymes. In this sense, three single nucleotide polymorphisms (SNP) in the XPD gene (in codons 156, 312, and 751) are related with different DNA repair capacity[5-7]. Thus, variants generated by amino acids can change Asp312Asn (exon 10, G > A substitution) and Lys751Gln (exon 23 A > C substitution) located in conserved regions of the XPD protein, and are associated with lower efficiency of damage repair and significantly higher background frequency of apoptotic cells in irradiated lymphocytes[8].
Although gastrectomy is the only potentially curative treatment in gastric-cancer patients, the overall survival results remain unsatisfactory. The main factor accounting for high mortality is relapse after surgical resection. During the past few decades, the principle of combined treatment modality has been developed and applied in various solid tumors including gastric cancer. In order to prevent recurrence and increase the cure rate of gastric-cancer after surgery, multiple studies using variable treatment modalities have been undertaken. One of the landmark studies reported that chemo-radiotherapy can significantly improve survival after resection of stage IB to stage IV gastric cancers[9]. Chemo-radiotherapy has been increasingly recognized as a standard of care since then[10,11]. However, whether adjuvant chemo-radiotherapy can prolong the survival of patients with extensive lymph node dissection remains debatable. In this paper, we evaluated the potential role of XPD codon 751 variant in the outcome of 44 patients with resected gastric cancer after chemo-radiotherapy.
Characteristics of the patients are listed in Table 1. From October 1992 to January 1999, 44 patients consisting of 32 men (73%) and 12 women (27%) with a median age of 60 years (range: 33-77 years) with diagnosis of gastric cancer after having undergone gastrectomy were treated with radiation plus 5-fluorouracil (5-FU) and leucovorin. Thirty-two percent of the patients had stageI-IIand 68% stage III-IV gastric cancer at the time of diagnosis (Table 1). The median follow-up time was 46.9 mo (range: 5.33 to 124.06 mo). The median time to progression was 37.5 mo (range: 2.3 to 95.4 mo). This study was conducted in Navarra Hospital Center, and informed consent was obtained from all the patients for using their tissues. The clinicopathological information of each subject was obtained from the tumor registry at Navarra Hospital.
| Baseline factors | n (%) ) |
| Sex | |
| Male | 32 (73) |
| Female | 12 (27) |
| Age (yr) | |
| ≤ 50 | 16 (36) |
| 51-64 | 13 (30) |
| ≥ 65 | 15 (34) |
| Median (Range) | 60 (33-77) |
| Stage at diagnostic | |
| I-II | 14 (32) |
| IIIA + IIIB | 21 (48) |
| IV | 9 (20) |
| Histotype | |
| Intestinal | 9 (20) |
| Diffuse | 35 (80) |
| Grading | |
| I | 3 (7) |
| II | 8 (18) |
| III | 33 (75) |
| Tumor location | |
| Cardia | 9 (21) |
| Fundus | 9 (21) |
| Body | 10 (24) |
| Antrum | 14 (33) |
| Gastrectomy | |
| R0 | 35 (80) |
| R1 | 8 (18) |
| R2 | 1 (2) |
| XPD 751 | |
| Lys/Lys | 19 (51) |
| Lys/Gln | 14 (38) |
| Gln/Gln | 4 (11) |
| Unknown | 7 |
The patients received postoperative treatment with 5-FU plus leucovorin and local radiation 20-40 d after gastrectomy. This chemotherapy regimen developed by the North Central Cancer Treatment Group[12] was used before and after radiation. Chemotherapy (fluorouracil, 425 mg/m2 per day, and leucovorin, 20 mg/m2 per day) was initiated on d 1 followed by chemo-radiotherapy on d 28 after the initial cycle of chemotherapy. Chemo-radiotherapy consisted of 4500 cGy of radiation at 180 cGy/d, 5 d/wk for 5 wk, with 5-FU (400 mg/m2 per day) and leucovorin (20 mg/m2 per day) on the first four and the last three days of radiotherapy. One month after radiotherapy, two 5-d cycles of 5-FU (425 mg/m2 per day) plus leucovorin (20 mg/m2 per day) were given every other month.
The 4500 cGy of radiation was delivered in 25 fractions (5 d/wk) to the tumor bed, regional nodes and 2 cm beyond the proximal and distal margins of resection. The adjuvant treatment was performed as previously described[9].
Surgical specimens (paraffin blocks) were cut into 5 μm-thick sections for molecular analysis.
Three tissue sections were transferred into a micro centrifuge tube and 1.2 mL of xylene was added. After centrifugation at 14 000 r/min for 5 min at room temperature (RT), the supernatant was removed. Subsequently, the tissue samples were washed in 1.2 mL of 96 mL/L ethanol. After centrifugation at 14 000 r/min for 5 min at RT, the supernatant was discarded. The washing procedure was repeated another time. The samples were dried for 3-5 min, in a vacuum pump (any letter between were and dried).
DNA was extracted using the QIAamp DNA Mini Kit (Qiagen-IZASA- Barcelona Spain). The deparaffinized samples were re-suspended in 180 μL of ATL buffer plus 20 μL of proteinase K and incubated overnight at 56°C. The samples were processed according to the provided protocol. The purified DNA was finally eluted in a total volume of 150 μL. DNA yield was quantified by NanoDrop 3.0.0 (Nucliber, Wilmington, Delaware USA).
About 7-509 mg/L of DNA was extracted and 45-200 ng of genomic DNA was used as template. PCR/RFLP-based assays were performed as described previously[5] with slight modifications. The PCR product (20 μL) was digested with 25 units of Pst I enzyme (New England BioLabs, -IZASA, S.A, Barcelona-, Spain) in 50 μL reaction mixture for 1 h. The digestion product was visualized in 3% agarose gel (Pronadisa, Madrid, Spain). The wild-type homozygote was defined by 104- and 220-bp banding patterns; the heterozygote by 63-, 104-, 157-, and 220-bp fragments; and variant homozygote by 63-, 104-, and 157-bp fragments. Some samples were noted as “unknown” because they could not be amplified due to a relatively frequent occurrence of PCR inhibitory substances in samples prepared with this DNA extraction method (Table 1). Furthermore, three DNA samples corresponding to each genotype selected by direct sequencing were used. The results were in concordance with RFLP genotyping.
This study was designed to analyze the role of XPD polymorphism in the prediction of relapse. Relapse free-survival (RFS) was defined as the time from the start of chemotherapy to the first evidence of disease progression. RFS was calculated using the Kaplan-Meier method. Contingency tables and chi-square test (χ2) were used to summarize the association of relapse with XPD polymorphism. All P values were two-sided. Cox proportional hazards models were also used to evaluate the different variables considered. All statistical tests were conducted by SPSS software 11.0 version for Windows (SPSS, Inc. Chicago). P ≤ 0.05 was considered statistically significant.
Thirty-seven patients were evaluated for the Lys751Gln polymorphism. Seven patient were not assessed due to poor sample extraction quality. Fifty-one percent (19 of 37) of the patients were homozygous for the Lys/Lys genotype, 38% (14/37) heterozygous for Lys/Gln, and 11% (4 of 37) homozygous for the glutamine variant (Table 1). The 751Gln allele frequency was 0.33, similar to that observed in other studies[5,7].
Sixteen patients (36%) developed grade III-IV toxicity. Hematological toxicity was found in 62% (10/16) patients compared with gastrointestinal and other toxicity in19% (3/16) patients (Table 2). However, there was no association between XPD genotype and toxicity (data not shown).
| Toxicity grade III-IV | n (%) |
| No | 28 (64) |
| Yes | 16 (36) |
| Toxicity type | |
| Hematological | 10 (62) |
| Digestive | 3 (19) |
| Others | 3 (19) |
Gastrectomy with D1 lymphadenectomy was performed in all patients. Of the 44 patients, 20 (45.5%) had relapse and 17 (85%) of them died. The first relapse site was local in 5, distant in 10, and both local and distant in 5 (Table 3).
| Relapse | n (%) |
| No | 24 (54.5) |
| Yes | 20 (45.5) |
| Relapse sites | |
| Local | 5 (25) |
| Distant | 10 (50) |
| Local + Distant | 5 (25) |
Statistical analysis showed that 75% (12 of 16) of relapse patients showed Lys/Lys genotype more frequently (P = 0.042, Table 4).
| Relapse | XPD | Stage | |||
| Lys | Lys/Gln | Gln/Gln | I+ II | IIIA + IIIB + IV | |
| No | 7 | 11 | 3 | 11 | 13 |
| Yes | 12 | 3 | 1 | 3 | 17 |
| P | 0.042 | 0.050 | |||
Other clinico-pathological features such as histology, grading and localization were not significant (data not shown).
The relative risk of progression (with Lys/Lys genotype used as the reference) was 0.35 (95% CI: 0.11-1.09) for patients carrying the Lys/Gln genotype and 0.25 (95% CI: 0.03-1.96) for the Gln/Gln group (Table 5).
| XPD | RFS (median) | Hazard Ratio | 95% CI |
| Lys/Lys | 11.79 | 1.0 | Reference |
| Lys/Gln | 14.33 | 0.35 | 0.11 to 1.09 |
| Gln/Gln | 23.33 | 0.25 | 0.03 to 1.96 |
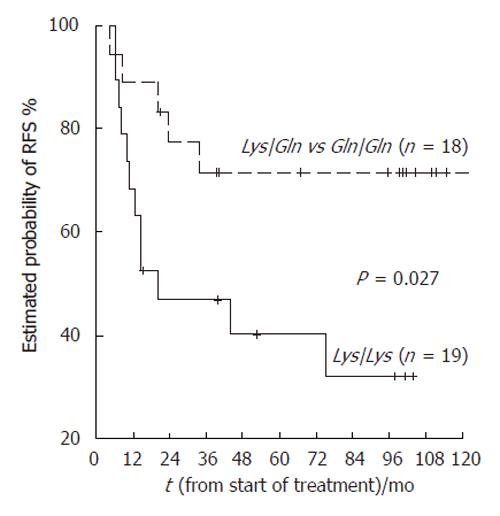
Only XPD genotype and stage to diagnosis showed a significant relation with cancer relapse and were evaluated as potential predictors of RFS by Cox test. We used the combined genotypes LysGln/GlnGln as reference in these analyses and found that the Lys/Lys genotype was more strongly associated with disease progression (P = 0.036) than the combined LysGln/GlnGln genotype (adjusted HR: 3.07, 95% CI: 1.07-8.78) (Table 6, Figure 1).
| XPD | RFS (median) | Hazard Ratio | 95% CI | P |
| Lys/Lys | 11.79 | 3.07 | 1.07 to 8.78 | 0.036 |
| Lys/Gln + Gln/Gln | 20.26 | 1.0 | Reference |
Most cases of gastric cancer are diagnosed at an advanced stage with poor prognosis. Surgery remains the only potentially curative treatment, but it is associated with a high rate of local recurrence and distant metastases. When irradiation is combined with surgical resection for all or a majority of tumors, both survival and local control appear to be better than those for the unresected patients[13]. Epidemiological studies have shown that Gln variant at XPD-751 polymorphism is partiality able to repair DNA in lung cancer[7,14]. In this sense, cancer treatment is to cause DNA damage and tumor cell apoptosis. Gln variant seems to show a lower ability to repair DNA damage and cancer patients are more sensitive to chemo-radiotherapy. In contrast, the presence of allele Lys represents a positive effect in different DNA repair pathways and a possible poor response to cancer treatment. In this study, we found a significant relationship between clinical response to chemo-radiotherapy and the XPD Lys751Gln polymorphism. Patients with the Lys/Lys genotype were more likely to have relapse compared to those with the combined Lys/Gln and Gln/Gln genotype.
Scientific publications are available but with diverging results in the XPD751 polymorphism and cancer risk or treatment efficiency[3,5-7]. Thus, decreased DNA repair has been associated with Lys allele[6]. Epidemiological study has reported association of this allele with a higher risk of basal cell carcinoma[5] even though no significant relationship between this polymorphic gene and DNA repair proficiency has been reported[15,16]. Spitz et al[7] and other research group[17] reported a suboptimal DNA repair capacity (DRC) particularly in subjects who were homozygous for 751Gln alleles. Moreover, a recently study has found a tendency toward a higher background frequency of apoptotic cells in irradiated lymphocytes carrying variant homozygote Gln at XPD codon 751[8]. Our results are in agreement with these studies, suggesting that heterozygote and homozygote Gln alleles may therefore increase their clinical response to suboptimal DNA damage repair after treatment.
Repair of DNA damage is a complex process[1,18,19]. Functional protein of XPD could be affected by a structural change (Lys → Gln) from a basic to a polar amino acid change located at about 50-base upstream from the poly (A) signal[5]. Moreover, close proximity in the genome between XPD and other polymorphic DNA repair genes such as X-ray repair cross complementing (XRCC1) and excision repair cross complementing (ERCC1) may influence the interaction between them leading to different or synergic DNA repair capability. A recent molecular epidemiology study has genotyped 44 SNPs in 20 genes involved in four DNA damage repair pathways, showing that homozygous Gln751Gln and other gene variants genotyped are highly associated with lung cancer risk[20]. In the same way, variant Gln allele and some nucleotide and base excision repair (NER and BER, respectively) variant gene members are associated with increased level of polycyclic aromatic hydrocarbon-DNA adducts[15,21].
In conclusion, XPD Lys751Gln polymorphism may be an important marker of genotoxicity that can predict the clinical outcome of resected gastric cancer treated with chemo-radiotherapy.
S- Editor Pan BR L- Editor Wang XL E- Editor Bai SH
| 1. | Sancar A. DNA repair in humans. Annu Rev Genet. 1995;29:69-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 145] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Flejter WL, McDaniel LD, Johns D, Friedberg EC, Schultz RA. Correction of xeroderma pigmentosum complementation group D mutant cell phenotypes by chromosome and gene transfer: involvement of the human ERCC2 DNA repair gene. Proc Natl Acad Sci USA. 1992;89:261-265. [RCA] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 111] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S, Lenz HJ. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344-354. [PubMed] |
| 4. | Takayama K, Salazar EP, Broughton BC, Lehmann AR, Sarasin A, Thompson LH, Weber CA. Defects in the DNA repair and transcription gene ERCC2(XPD) in trichothiodystrophy. Am J Hum Genet. 1996;58:263-270. [PubMed] |
| 5. | Dybdahl M, Vogel U, Frentz G, Wallin H, Nexø BA. Polymorphisms in the DNA repair gene XPD: correlations with risk and age at onset of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev. 1999;8:77-81. [PubMed] |
| 6. | Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, Bell DA. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21:551-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 316] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 7. | Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61:1354-1357. [PubMed] |
| 8. | Rzeszowska-Wolny J, Polanska J, Pietrowska M, Palyvoda O, Jaworska J, Butkiewicz D, Hancock R. Influence of polymorphisms in DNA repair genes XPD, XRCC1 and MGMT on DNA damage induced by gamma radiation and its repair in lymphocytes in vitro. Radiat Res. 2005;164:132-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA, Gunderson LL, Jessup JM. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2465] [Cited by in RCA: 2437] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 10. | Macdonald JS. Adjuvant therapy for gastric cancer. Semin Oncol. 2003;30:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Macdonald JS. Clinical overview: adjuvant therapy of gastrointestinal cancer. Cancer Chemother Pharmacol. 2004;54 Suppl 1:S4-S11. [PubMed] |
| 12. | Poon MA, O'Connell MJ, Moertel CG, Wieand HS, Cullinan SA, Everson LK, Krook JE, Mailliard JA, Laurie JA, Tschetter LK. Biochemical modulation of fluorouracil: evidence of significant improvement of survival and quality of life in patients with advanced colorectal carcinoma. J Clin Oncol. 1989;7:1407-1418. [PubMed] |
| 13. | Carrato A, Gallego-Plazas J, Guillen-Ponce C. Adjuvant therapy of resected gastric cancer is necessary. Semin Oncol. 2005;32:S105-S108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Palli D, Russo A, Masala G, Saieva C, Guarrera S, Carturan S, Munnia A, Matullo G, Peluso M. DNA adduct levels and DNA repair polymorphisms in traffic-exposed workers and a general population sample. Int J Cancer. 2001;94:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 94] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 15. | Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, Ashok TD, Mark EJ, Wain JC, Christiani DC, Kelsey KT. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21:965-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 325] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 16. | Moller P, Knudsen LE, Frentz G, Dybdahl M, Wallin H, Nexo BA. Seasonal variation of DNA damage and repair in patients with non-melanoma skin cancer and referents with and without psoriasis. Mutat Res. 1998;407:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509:165-174. [RCA] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Miller MC 3rd, Mohrenweiser HW, Bell DA. Genetic variability in susceptibility and response to toxicants. Toxicol Lett. 2001;120:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 102] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 337] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 20. | Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 306] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 21. | Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 346] [Article Influence: 14.4] [Reference Citation Analysis (0)] |