Published online Aug 7, 2006. doi: 10.3748/wjg.v12.i29.4673
Revised: September 28, 2005
Accepted: January 15, 2006
Published online: August 7, 2006
AIM: Hepatitis B virus protein X (HBx) has been shown to be weakly oncogenic in vitro. The transforming activities of HBx have been linked with the inhibition of several functions of the tumor suppressor p53. We have studied whether HBx may have different effects on p53 depending on the cell type.
METHODS: We used the human hepatoma cell line HepG2 and the immortalized murine hepatocyte line AML12 and analyzed stably transfected clones which expressed physiological amounts of HBx. P53 was induced by UV irradiation.
RESULTS: The p53 induction by UV irradiation was unaffected by stable expression of HBx. However, the expression of the cyclin kinase inhibitor p21waf/cip/sdi which gets activated by p53 was affected in the HBx transformed cell line AML12-HBx9, but not in HepG2. In AML-HBx9 cells, p21waf/cip/sdi-protein expression and p21waf/cip/sdi transcription were deregulated. Furthermore, the process of apoptosis was affected in opposite ways in the two cell lines investigated. While stable expression of HBx enhanced apoptosis induced by UV irradiation in HepG2-cells, apoptosis was decreased in HBx transformed AML12-HBx9. P53 repressed transcription from the HBV enhancer I, when expressed from expression vectors or after induction of endogenous p53 by UV irradiation. Repression by endogenous p53 was partially reversible by stably expressed HBx in both cell lines.
CONCLUSION: Stable expression of HBx leads to deregulation of apoptosis induced by UV irradiation depending on the cell line used. In an immortalized hepatocyte line HBx acted anti-apoptotic whereas expression in a carcinoma derived hepatocyte line HBx enhanced apoptosis.
- Citation: Fiedler N, Quant E, Fink L, Sun J, Schuster R, Gerlich WH, Schaefer S. Differential effects on apoptosis induction in hepatocyte lines by stable expression of hepatitis B virus X protein. World J Gastroenterol 2006; 12(29): 4673-4682
- URL: https://www.wjgnet.com/1007-9327/full/v12/i29/4673.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i29.4673
The epidemiological association between chronic HBV infection and the development of hepatocellular carcinoma is well established[1]. However, the molecular mechanism of transformation by HBV is unsolved. Among many potential factors, a viral regulatory protein named Hepatitis B virus protein X (HBx) is believed to contribute to oncogenesis in conjunction with other mechanisms. Stable expression of HBx showed a weak transforming effect in cell lines of liver origin[2-5], in murine fibroblasts[6,7] and in rat fibroblasts[8-12]. Neither the molecular actions set in motion by HBx during the process of transformation, nor the role of HBx for HBV replication are well understood. HBx is dispensable for replication in vitro[13], but under certain conditions HBx can enhance replication in vitro[14-16]. In the woodchuck, a related virus of the same family of viruses, the hepadnaviridae[17], the analog of HBx is essential for the establishment of infection[18,19]. A plethora of in vitro activities and interactions with cellular partners by HBx have been reported[22,23].
In the development of cancer, the delicate balance between cell proliferation and programmed cell death is often disturbed[24]. Several functions of HBx are linked to transformation: HBx transactivates the transcription of cellular proto-oncogenes[25-27], and is able to override cell cycle check points[28,29]. As the oncoproteins of many tumor viruses influence the course of apoptosis in order to transform their target cells[30-32], several reports described the influence of stable and transient transfection of HBV products on apoptosis. Transfection of replication competent constructs of HBV[33] and the closely related hepadnavirus woodchuck hepatitis virus[34] enhanced apoptosis induction. This pro-apoptotic activity of hepadnavirus genomes was dependent on the integrity of the HBx-ORF[33,34]. Several groups described a pro-apoptotic activity of HBx after transient and stable transfection[33,34-48] whereas other groups described anti-apoptotic effects of HBx[49-59].
The cellular tumor suppressor protein p53 is one of the key players in the induction of apoptosis after genotoxic events[60,61]. Thus, besides influencing programmed cell death the transforming proteins of DNA tumor viruses target and inactivate p53 in quite diverse and elaborate ways[31,32]. Several lines of evidence connect HBx with a disturbance of p53 functions. HBx binds to p53 in vitro[62,63], the intracellular distribution of p53 appears to be affected by HBx[4,64,65], p53-dependent and independent DNA repair functions are affected by HBx[66-70]. Furthermore, HBx relieves the repression of transcription by the tumor suppressor p53 in transient reporter gene assays[71-73], and influences the expression of the p53 induced cyclin kinase inhibitor p21waf/cip/sdi[74].
However, most of the experiments in these reports were done in transient transfection or even cell free in vitro systems. Because the number of HBx molecules in naturally infected cells is assumed to be low[75-77] and transient transfections of HBx expression constructs yield very high amounts of HBx protein, we established cell lines stably expressing low amounts of HBx. Next, we studied the effect of HBx expression on several functions of p53 induced by stimuli that were designed to yield physiological p53 amounts in our cell lines. Using these bona fide natural conditions we were able to find that even low levels of HBx altered or even antagonized p53 functions.
AML12: The highly differentiated murine hepatocyte line AML12[78] derived from mice transgenic for TGFα can be transformed by HBx[3]. AML12 clones stably transfected with and transformed by HBx and the control cell lines stably transfected with pSV2neo were obtained from N. Fausto. AML12 clones were cultured in Dulbecco’s MEM/Nutrient Mix F12 supplemented with 10% FCS, 0.1 μM Dexamethason, and ITS (Insulin, Transferrin, and Selene, 5 μg/mL each).
HepG2: The differentiated but already transformed human hepatoblastoma line HepG2[79] was cultivated in Mixed Medium/10% FCS.
For CAT assays, the cells per well were transfected with Lipofectamine (Life Technologies, Karlsruhe). Detection of CAT protein was done with the CAT-ELISA Kit (Roche, Mannheim) according to the instructions of the manufacturer.
All cell lines were plated out one day before irradiation. Directly before irradiation, the medium was removed and the cultures were washed with PBS. UV irradiation was done in the UV-Stratalinker (Stratagene, Leimen, Germany) at 254 nm. The cells were exposed to UV after removing the cover of the dishes.
For induction of apoptosis, 1000 cells were plated in triplicates on culture dishes with a diameter of 95 mm. After irradiation with the indicated energy in the stratalinker, the cell cultures were grown for 5-15 d, until colonies visible by eye were detected. After washing with PBS, the colonies were stained and fixed with 1% crystal violet in 20% methanol for 5 min. After washing with H2O, the surviving colonies were counted. Each experiment was done in triplicate and repeated at least thrice.
For the lysis of cells, 450 μL of ice cold lysis buffer were added to each well of a six-well plate for 30 min on ice. The lysis was based on RIPA-buffer (0.01 mol/L Tris-HCl, pH 7.4; 0.15 mol/L NaCl; 1% (w/v) sodium desoxycholate; 0.1% (w/v) SDS; 1% Triton X-100) supplemented with Leupeptin and Aprotinin. To digest disturbing chromosomal DNA 250 U/mL Benzonase (Novagen, Merck, Darmstadt) was added to the lysis buffer. After the incubation period, the lysate was cleared by centrifugation at 20 000 g for 5 min. The protein content of the lysates was determined using the BCA assay and equal amounts of protein (20-50 μg) were separated on an SDS gel. After blotting and incubation with the appropriate primary and secondary antibodies, the membranes were developed using Chemiluminescence Blotting Substrate from Boehringer Mannheim, Germany according to the manufacturer’s instructions.
The following antibodies were used for the detection of the indicated proteins: murine p53: AB7 (biotinylated polyclonal sheep antibodies from Oncogene Science, via Dianova, Hamburg, Germany); human p53: mAb PAb1801 (Pharmingen, Hamburg, Germany); murine p21: mAb AB4 (OP76 Oncogene, via Dianova, Hamburg, Germany); human p21: p21 (187) (sc-817), St Cruz Biotechnology.
As secondary antibody, we used POD-labeled anti-murine IgG from donkey (Jackson ImmunoResearch, via Dianova, Hamburg, Germany) or POD-strepatividin (Calbiochem, Bad Soden, Germany) for murine p53.
For Northern blot, total cellular RNA was extracted from the cells using TRIzol (Gibco). Twenty micrograms of DNase I digested RNA was separated on a glyoxal agarose gel (1% in 10 mmol/L Na2HPO4) and blotted onto positively charged nylon membrane. The blot was hybridized with a 32P labeled p21-specific probe derived from pCMVp21 (murine).
For RPA, an RNA probe complementary to human p21 was transcribed with Maxiscript, Ambion from the construct pCMVsdi[80] according to the instructions of the manufacturer. As an internal control, we used actin transcribed from pTRI-β-Actin (Ambion, Austin, TX, USA).
The RNA probe was purified by electrophoresis in 5% acrylamide/8 mol/L urea gel. The probe (0.5-2 μg) was labeled with biotin using the BrightStarTM Psoralen-Biotin kit (Ambion, Austin, TX, USA).
The RPA was performed with the HybSpeedTM RPA kit from (Ambion, Austin, TX, USA). After digestion of the samples as described in the manual of the kit, the samples were run on 5% PAGE/8 mol/L urea in TBE and blotted onto a positively charged nylon membrane (Amersham, Braunschweig). Signal detection was performed with streptavidin labeled with alkaline phosphatase using the BrightStarTM BioDetectTM kit from Ambion, Austin, TX, USA.
For RT-PCR, 2 μg of total cellular RNA extracted with Qiagen RNeasy was digested with DNase I for 45 min. One microgram of DNase I digested RNA was reverse transcribed with superscript (Gibco). One quarter of the reverse transcribed RNA was amplified with Taq polymerase (1´ 94°C, 1´ 54°C, 1´ 72°C: × 35 cycles) using HBx-specific primers, which had been used for cloning the complete HBx ORF into pRcCMV.
pRcCMVX: The complete HBx ORF of an HBV genotype A genome (serotype adw2; isolate 991; EMBL accession no.: X51970) was cloned with the help of PCR into the EcoRI and XbaI-site of pCDNA3, Invitrogen, and controlled by sequencing.
pRcCMVXM: Identical to pRcCMVX except for two stop codons introduced by a G to A exchange of nt 1443 (GAA→TAA)[2] and a C to A exchange of nt 1684 (TCA→TAA). Thus, translation of full-length HBx is stopped by exchange of nt 1443 after 23 aa and expression of presumed small HBx proteins is made impossible by exchange of nt 1684, which introduces a stop directly after the third start codon in frame in the HBx-ORF.
PS9: HBV enhancer I (nt 1040-1374 from EMBL accession no.: X51970) was cloned into pBLCAT3[81].
pC53SN3: Expression vector for wt-p53[82]. P53 is expressed from the CMV enhancer in the vector pCMV.
pcDNA3p21 (murine): Murine p21 was cloned by RT-PCR with primers waf-AS and waf-S[33] from total cellular RNA derived from AML12 cells. After EcoRI digestion, the amplicon was cloned into pCDNA3. Sequencing gave complete identity with murine p21 from GenBank.
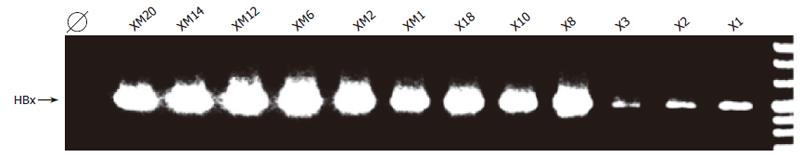
HepG2 cells-a completely transformed hepatoma cell line- were stably transfected with the HBx-expression construct pRcCMVX. As a negative control, HepG2 cells were transfected with the isogenic HBx protein expression-negative construct pRcCMVXM. Clones obtained after G418 treatment were analyzed for the expression of HBx-RNA expression by RT-PCR (Figure 1). 11/13 wt-HBx transfected and 11/11 HBxM transfected clones expressed HBx RNA detectable by RT-PCR. While all HBxM transfected clones expressed similar amounts of HBx RNA as judged by RT-PCR, it appeared as if HBx RNA expression in wt-HBx transfected clones varied significantly (Figure 1). This reminds of the findings in stably HBx transfected clones of rat fibroblast origin, where HBx expression was downregulated in most clones[33].
Clone HepG2X8 expressing wt-HBx and control clone HepG2 XM2 transfected with the HBx-expression minus construct pRcCMVXM expressed similar amounts of HBx-RNA and were chosen to be studied in detail.
In comparison, we analyzed the immortalized murine hepatocyte line AML12[78] and the AML12-clone HBx9 transformed by HBx[3].
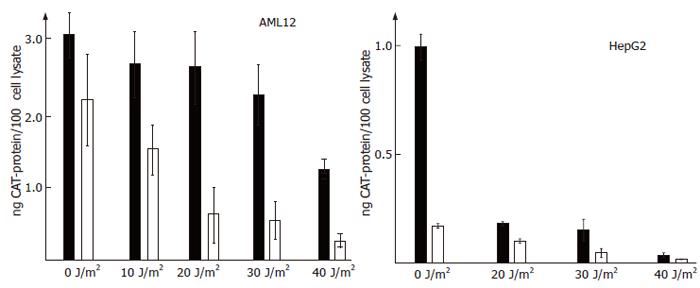
One well-characterized function of HBx is its potential to transactivate virtually any promoter or enhancer. We were unable to detect HBx protein in stably transfected AML12 and HepG2 clones (data not shown and Ref. [3])-a problem encountered by many other groups. In order to show the presence of active HBx, we analyzed whether transcriptional activation by HBx could be detected in cell lines stably expressing HBx RNA. Indeed, the expression from HBV enhancer I was approximately sevenfold stronger in the HBx expressing cell line HepG2-X8 than in the control cell line HepG2-XM2 (Figure 2B). In AML12 cell transactivation by HBx was lower (Figure 2A).
To analyze the extent of apoptosis quantitatively, we investigated the survival of the cell lines under investigation after UV irradiation. Five hundred cells in triplicate were plated and irradiated with the dose indicated. Surviving colonies were counted after about 14 d. Table 1 shows that stable expression of HBx acts pro-apoptotic in HepG2-derived and anti-apoptotic in AML12-derived cell lines. The pro-apoptotic effect of HBx in HepG2-X8 was most obvious in cell lines irradiated with 20 J/m2. For example in one experiment 17.7% ± 8.0 cells of the control cell line HepG2-XM2 survived irradiation with 20 J/m2, whereas only 1.1% ± 0.6 of identically treated HBx expressing HepG2-X8 cells were able to grow out to visible colonies. AML12 cells were more sensitive against UV irradiation. Expression of HBx acted anti-apoptotic in the HBx expressing AML12 cell line and enhanced the survival rates to similar levels as in HepG2XM control cells.
| 0 J/m2 | 20 J/m2 | 50 J/m2 | |
| HepG2-XM2 | 100 ± 1.4 | 17.7 ± 8.0 | 1.0 ± 0.9 |
| HepG2-X8 | 100 ± 20.9 | 1.1 ± 0.6 | 0.2 ± 0.1 |
| HepG2-XM2 | 100 ± 11.8 | 15.8 ± 7.1 | 1.38 ± 1.5 |
| HepG2-X8 | 100 ± 20.3 | 2.7 ± 1.5 | 0.0 |
| HepG2-XM2 | 100 ± 15.6 | 13.5 ± 6.9 | 3.7 ± 0.8 |
| HepG2-X8 | 100 ± 7.4 | 6.8 ± 0.5 | 1.7 ± 1.2 |
| AML12 | 100 ± 5.1 | 4.3 ± 0.6 | 1.1 ± 1.0 |
| AML12-HBx9 | 100 ± 14.2 | 13.5 ± 8.9 | 6.0 ± 2.2 |
| AML12 | 100 ± 27.2 | 17.0 ± 6.3 | 0.6 ± 0.04 |
| AML12-HBx9 | 100 ± 3.8 | 28.0 ± 7.0 | 6.8 ± 2.4 |
| AML12 | 100 ± 4.4 | 11.0 ± 4.1 | 0.4 ± 0.5 |
| AML12-HBx9 | 100 ± 4.6 | 17 ± 5.1 | 3.4 ± 1.8 |
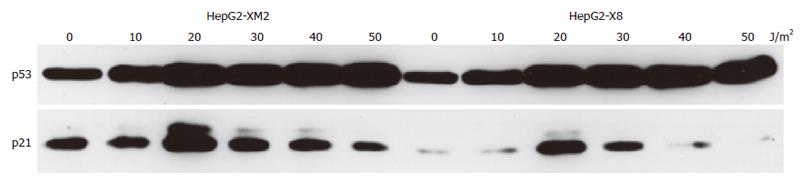
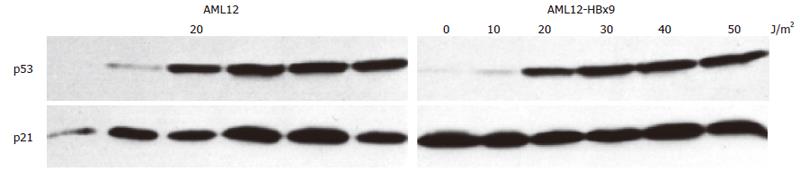
p53 is a central protein in the induction of apoptosis[83]. After many sublethal stimuli, a complex set of events takes place that results in an increased expression of p53 in the nucleus. Thus, we analyzed by immune blots of whole cell extracts whether HBx was able to modulate expression of p53 after UV irradiation of the cell lines. Irradiation with increasing doses (0-50 J/m2) of UV led to enhanced expression of p53 in control and HBx expressing cell lines (Figures 3 and 4). HBx had no effect on the induction of p53 in cell lines of AML12 and HepG2 origin. p53 expression was easily detectable after irradiation with 20 J/m2 and maximal expression was reached after irradiation with 40-50 J/m2 in the HepG2 and AML12 cell lines investigated (Figures 3 and 4). These data are in accordance with reports on p53 expression in HepG2-cells[84], human[85], and murine[85,86] fibroblasts and primary rat keratinocytes[87].
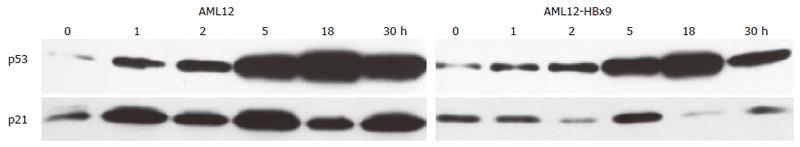
We also analyzed the kinetics of p53 induction after UV irradiation with 40 J/m2. p53 levels increased after 1 h to reach a maximum 18 h after irradiation. Thereafter, p53 expression decreased (Figure 5). Again, HBx had no influence on the kinetics of p53 induction after UV irradiation.
We next asked whether HBx may influence functions of p53. One extensively investigated activity of p53 is the ability to repress the transcription of many genes[88]. Thus, we analyzed whether expression of the reporter gene CAT from HBV enhancer I is repressed by p53 - or other factors activated after induction of programmed cell death - in vivo. Irradiation of HepG2 and AML12-cell lines with increasing doses of UV caused indeed a strong repression of expression from HBV enhancer I (Figures 2A and B). The repression was highest at 40 J/m2 (Figures 2A and B), the UV dose leading to maximum expression of p53 (Figures 2 and 3). However, a strong reduction of CAT-repression could already be observed at the lowest UV dose of 10 J/m2 administered. Interestingly, in the tumor cell line HepG2 (Figure 2B), UV irradiation caused a much stronger repression of expression from enhancer I than in AML12 cells (Figure 2A).
In HBx expressing cell lines, the UV-induced repression was partially counteracted. This anti-repressive activity of HBx was strongest in AML12-HBx9 and still significant in HepG2 cells (Figures 2A and B). In the AML-cell line HBx9 irradiated with 40 J/m2 the CAT-expression was 4.9-fold higher than in the parental cell line AML12 without HBx. Thus, HBx seems to be able to alleviate transcriptional repression by p53.
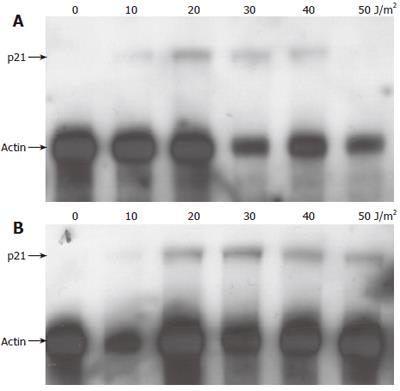
Because we observed that HBx influenced the extent of transcriptional repression by p53 we also wanted to analyze if HBx had any influence on transcriptional activation by p53. After UV irradiation, p53 is stabilized[89] and acts as a transcription factor by binding to the promoter of certain target genes leading to enhanced transcription from these promoters, for example of the cyclin kinase inhibitor p21waf/cip/sdi[90]. Thus, after UV irradiation, the expression of p21waf/cip/sdi is enhanced[86,91]. We therefore analyzed if stable expression of HBx influenced p53’s transcriptional activation of the endogenous p21waf/cip/sdi-promoter. UV irradiation of the cell lines with increasing doses of UV led to an increase of p21waf/cip/sdi protein expression (Figures 3 and 4). As reported by other groups, irradiation with high doses of 40-50 J/m2 UV again caused a decline of the induced p21waf/cip/sdiwaf protein expression[86,91]. An analysis of the temporal regulation of p21waf/cip/sdiwaf protein expression (Figure 5) after UV irradiation in AML12 cells showed an undulation of p21waf/cip/sdiwaf protein expression in the parental cell line and the HBx transformed clone 9. A similar phenomenon was described in the mouse liver after 70% partial hepatectomy[92]. However, these authors analyzed RNA expression and different time points. Interestingly, stable expression of HBx in HepG2 led to a lower expression of p21waf/cip/sdi-protein. However, in the HBx transformed cell line AML12-HBx9, the level of p21waf/cip/sdi-protein expression after UV irradiation changed from experiment to experiment and no reproducible data for p21 could be obtained, although p53-expression from identical blots (upper lane) gave reproducible results (Figures 4 and 5). It appeared as if the expression of p21waf/cip/sdi-protein was deregulated in a seemingly random way by HBx because p21waf/cip/sdi-protein expression was reproducible in the parental cell line (Figures 4 and 5).
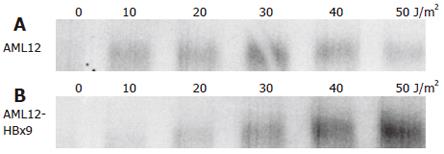
Butz et al had described an uncoupling of the regularly inducible p21waf/cip/sdi-RNA expression from the deregulated p21waf/cip/sdi-protein expression upon genotoxic stress in some cell lines[109]. Thus, we analyzed the induction of p21waf/cip/sdi-RNA in the HBx transfected cell lines. Indeed, analysis of p21waf/cip/sdi-RNA expression in the hepatocyte lines under investigation gave reproducible results. Irradiation with UV increased the expression of p21waf/cip/sdi-RNA in the HBx-expressing cell lines HepG2-X8 and the control cell lines HepG2-XM2 and AML12 to reach a peak at 20-30 J/m2 to decrease thereafter (Figures 6 and 7). Analysis of p21waf/cip/sdi-RNA expression in the HBx expressing clone AML12-HBx9 - in contrast to analysis of p21waf/cip/sdi-protein expression - gave reproducible results. However, no decline of p21waf/cip/sdi-RNA expression was observed at higher doses of UV irradiation as with the other three cell lines under investigation (Figure 7). Quite in contrast, in the HBx-expressing clone AML12-HBx9 the amount of p21waf/cip/sdi-RNA increased continuously even at the highest dose of UV irradiation of 50 UV J/m2. Thus, HBx strongly influenced the regulation of p21waf/cip/sdi expression in the HBx-transformed cell line AML12-HBx9.
One of the most important functions of p53 is the induction of programmed cell death in order to maintain the integrity of the genome by inhibiting the survival of cells with damaged DNA[93]. To analyze the integrity of p53 function in cell lines stably expressing HBx RNA, we investigated several p53 activities during induction of apoptosis by UV. Irradiation with 20 J/m2 quite uniformly induced massive cell death in all cell lines analyzed. The cell death was due to apoptosis because we observed nucleosomal DNA fragmentation in all cell lines under investigation (data not shown).
However, we observed that induction of apoptosis was enhanced in HBx expressing cell lines of HepG2-origin and reduced in HBx expressing cell lines of AML12-origin. These opposing effects of HBx are in agreement with the published results on the influence of HBx on programmed cell death. Several groups described a pro-apoptotic activity of HBx after transient and stable transfection[33,34-48] whereas other groups described anti-apoptotic effects of HBx[49-59].
We thus set out to study key events in the induction of apoptosis after UV irradiation. We investigated whether HBx had an effect on the induction of p53 in our cell lines. No evidence was found that HBx influenced the induction of p53 by UV. Neither p53’s expression by stabilization of p53 as reported for adenovirus E1A or the large T-Antigen of SV40[94] was increased, nor was p53’s expression decreased in the presence of HBx as described for E6 of the papilloma viruses[95]. Our results agree with reports from Refs. [85] and [97] who also did not find altered p53 induction or expression in HBV expressing HepG2 cells.
A widely accepted hypothesis assumes that a higher expression of the cyclin kinase inhibitor p21waf/cip/sdi after DNA damage gives an advantage for survival because cells arrest in G1 and gain time to repair DNA damage instead of succumbing to apoptosis[88]. DNA damage induced in cell lines with a targeted deletion of the p21waf/cip/sdi-gene led to programmed cell death, whereas the wt-cell line survived in cell cycle arrest[97]. After induction, p53 binds as a transcription factor to the promoter of p21waf/cip/sdi and increases its transcription[90]. Our results on induction of p21waf/cip/sdi-protein and RNA by UV irradiation in the control cell lines (AML12 and HepG2-XM2) are in accordance with data from literature[86,87,91]. Interestingly, the HBx-expressing clone HepG2-clone X8 expressed less p21waf/cip/sdi than the control. A decrease as well as an increase of p21waf/cip/sdi expression in response to HBV infection has been described in literature. In microarrays, an enhanced expression of p21waf/cip/sdi was seen in Hep3B and HepG2 lines stably expressing HBx[96,98] and in the HBV expressing cell line HepG2.2.15[99], decreased expression of p21waf/cip/sdi was seen in HBV associated HCC[100], in transiently transfected primary human hepatocytes[101] and in two liver samples from patients with chronic HBV infection[101].
The reason for the differential effect of HBx on the expression of p21waf/cip/sdi in AML12 and HepG2 cell lines is unknown. However, transduction of HBx into primary human hepatocytes and stably transformed hepatocyte lines had reciprocal effects on the expression of several target genes[101]. It appears possible that stable expression of HBx in the immortalized AML12 cell line mimics the situation found in acutely infected liver with deregulated and in some circumstances enhanced p21waf/cip/sdi expression[100], whereas stable expression of HBx in hepatoma cell line HepG2 with slightly decreased expression of p21waf/cip/sdi mirrors the situation found in HBV associated HCC[100], i.e. in the endpoint of hepatocarcinogenesis.
It appears possible that HBx transactivates the enhancer of p21waf/cip/sdi in AML12 cells directly, as has been reported for other cell lines[74,96]. In addition, HBx may enhance the physiological activation of p21waf/cip/sdi by p53. Haviv et al[102] have shown that HBx can drastically enhance p53 mediated transcriptional activation. In consequence, HBx expressing AML12 clones would express more p21waf/cip/sdi than controls. Furthermore, HBx may have a stronger effect on the activation of the transcription factor ets in AML12 cells than in HepG2 cells, as ets has been suggested to play a crucial role in the transactivation of the p21waf/cip/sdi enhancer by HBx[96]. These assumptions would be in agreement with the higher and deregulated expression of p21waf/cip/sdi-RNA in the HBx expressing AML12 cell line. An enhanced expression of p21waf/cip/sdi in the HBx transformed cell line AML12-HBx9 reminds of findings in HTLV-1 transformed cell lines[103,104]. Tax, the transforming protein of HTLV-1 virus[105] has many functional similarities with HBx[106]. The oncoprotein E7 of the papilloma viruses also enhances the expression of p21waf/cip/sdi[107,108]. As described for otherwise unrelated cell lines[109], we also observed that the expression of p21waf/cip/sdi-protein was deregulated and did not seem to correlate with p21waf/cip/sdi-RNA-expression in HBx-expressing cell line AML12-HBx9 (Figures 4, 5 and 7).
Our results indicate that HBx is able to partially relieve repression of expression from the HBV enhancer I by physiological amounts of p53 as observed after induction by UV irradiation (Figure 2). Other groups reported complete relief of transcriptional repression by p53 after transient transfection of HBx[71] or HBV-dimers[72] or in in vitro transcription assays using in vitro translated p53 and HBx proteins[73]. However, transient transfection of HBx expression constructs leads to high expression of HBx, whereas stable expression of HBx as in our experiments only leads to low level expression of HBx that may be more comparable to the natural situation in tissue of chronically infected individuals where detection of HBx is rather difficult because of its low level of expression[75-77]. Moreover, in transient assays using expression vectors for p53 and HBx, a complete relief of transcriptional repression by p53 was only seen when 100-fold more HBx expression vector than p53 was used[71]. When equal amounts of expression constructs were used, HBx was not able to relieve transcriptional repression by p53[71,110]. Our results indicate that stable expression of HBx in hepatocyte lines antagonizes functions of physiological doses of p53 induced by UV and confirm the results of transient transfections[71,72].
In summary, we found several p53 linked activities that were altered in cell lines stably expressing HBx. By use of conditions which closely match the natural situation, we were able to show that HBx is able to alleviate some of the p53 induced effects taking place after the induction of apoptosis by UV irradiation. The differences we observe between the HBx expressing immortalized hepatocyte line AML12 and the hepatoma cell line HepG2 are due to a differential activation of intracellular signal transduction pathways - as found in inducible HBx expressing AML12-clones[5,111] - and will be the subject of further investigations.
This work was performed in partial fulfillment of the requirements for the Ph.D. thesis of N.F. and for the MD. thesis of E.Q.
We thank Dr. N. Fausto for the AML12 and AML12-HB9 cell line. We thank Dr. J. R. Smith, Baylor College of Medicine for the gift of the construct pCMVsdi and Dr. B. Vogelstein for pC53SN3.
S- Editor Wang J L- Editor Christy M E- Editor Liu WF
| 1. | Bréchot C, Jaffredo F, Lagorce D, Gerken G, Meyer zum Büschenfelde K, Papakonstontinou A, Hadziyannis S, Romeo R, Colombo M, Rodes J. Impact of HBV, HCV and GBV-C/HGV on hepatocellular carcinomas in Europe: results of a European concerted action. J Hepatol. 1998;29:173-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Seifer M, Höhne M, Schaefer S, Gerlich WH. In vitro tumorigenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13 Suppl 4:S61-S65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 58] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Oguey D, Dumenco LL, Pierce RH, Fausto N. Analysis of the tumorigenicity of the X gene of hepatitis B virus in a nontransformed hepatocyte cell line and the effects of cotransfection with a murine p53 mutant equivalent to human codon 249. Hepatology. 1996;24:1024-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Schaefer S, Seifer M, Grimmsmann T, Fink L, Wenderhold S, Höhne MW, Gerlich WH. Properties of tumour suppressor p53 in murine hepatocyte lines transformed by hepatitis B virus X protein. J Gen Virol. 1998;79:767-777. [PubMed] |
| 5. | Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327-2336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Shirakata Y, Kawada M, Fujiki Y, Sano H, Oda M, Yaginuma K, Kobayashi M, Koike K. The X gene of hepatitis B virus induced growth stimulation and tumorigenic transformation of mouse NIH3T3 cells. Jpn J Cancer Res. 1989;80:617-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 134] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 7. | Seifer M, Höhne M, Schaefer S, Gerlich WH. Malignant transformation of immortalized cells by hepatitis B virus DNA. Viral Hepatitis-1990. New York: Williams and Wilkins 1991; 586-588. |
| 8. | Gottlob K, Pagano S, Levrero M, Graessmann A. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 1998;58:3566-3570. [PubMed] |
| 9. | Buendia MA, Paterlini P, Tiollais P, Bréchot C. Hepatocellular carcinoma: molecular aspects. Viral hepatitis. 2nd ed. London: Churchill Livingstone 1998; 179-200. |
| 10. | Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol. 2000;10:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 222] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 11. | Schaefer S. Hepatitis B Virus in Experimental Carcinogenesis. Viruses, cell transformation and cancer. Greenwich: Elsevier 2001; 193-228. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 12. | Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093-5107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 374] [Article Influence: 17.0] [Reference Citation Analysis (1)] |
| 13. | Blum HE, Zhang ZS, Galun E, von Weizsäcker F, Garner B, Liang TJ, Wands JR. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223-1227. [PubMed] |
| 14. | Bouchard MJ, Puro RJ, Wang L, Schneider RJ. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J Virol. 2003;77:7713-7719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Zhang Z, Protzer U, Hu Z, Jacob J, Liang TJ. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J Virol. 2004;78:4566-4572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Burrell CJ, Chisari FV, Gerlich WH, Gowans EJ, Howard CR, Kann M. Hepadnaviridae. Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press 2000; 325-334. |
| 17. | Chen HS, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218-1226. [PubMed] |
| 18. | Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026-2030. [PubMed] |
| 19. | Yen TS. Hepadnaviral X Protein: Review of Recent Progress. J Biomed Sci. 1996;3:20-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 123] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Madden CR, Slagle BL. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis Markers. 2001;17:153-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 22. | Waris G, Siddiqui A. Regulatory mechanisms of viral hepatitis B and C. J Biosci. 2003;28:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 2.7] [Reference Citation Analysis (1)] |
| 23. | Kanzler S, Galle PR. Apoptosis and the liver. Semin Cancer Biol. 2000;10:173-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Avantaggiati ML, Natoli G, Balsano C, Chirillo P, Artini M, De Marzio E, Collepardo D, Levrero M. The hepatitis B virus (HBV) pX transactivates the c-fos promoter through multiple cis-acting elements. Oncogene. 1993;8:1567-1574. [PubMed] |
| 25. | Kekulé AS, Lauer U, Weiss L, Luber B, Hofschneider PH. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 277] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 26. | Twu JS, Lai MY, Chen DS, Robinson WS. Activation of protooncogene c-jun by the X protein of hepatitis B virus. Virology. 1993;192:346-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 89] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Koike K, Moriya K, Yotsuyanagi H, Iino S, Kurokawa K. Induction of cell cycle progression by hepatitis B virus HBx gene expression in quiescent mouse fibroblasts. J Clin Invest. 1994;94:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 106] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Benn J, Schneider RJ. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215-11219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 236] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 29. | Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 574] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 30. | Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 218] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 31. | Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1292] [Cited by in RCA: 1313] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 32. | Schuster R, Gerlich WH, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene. 2000;19:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Su F, Schneider RJ. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1997;94:8744-8749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 251] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 34. | Chirillo P, Pagano S, Natoli G, Puri PL, Burgio VL, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162-8167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 157] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 35. | Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 164] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 36. | Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon ML, Tiollais P, Buendia MA. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115-2123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Bergametti F, Prigent S, Luber B, Benoit A, Tiollais P, Sarasin A, Transy C. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene. 1999;18:2860-2871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 38. | Shintani Y, Yotsuyanagi H, Moriya K, Fujie H, Tsutsumi T, Kanegae Y, Kimura S, Saito I, Koike K. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol. 1999;80:3257-3265. [PubMed] |
| 39. | Chang SF, Netter HJ, Hildt E, Schuster R, Schaefer S, Hsu YC, Rang A, Will H. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J Virol. 2001;75:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Schuster R, Hildt E, Chang SF, Terradillos O, Pollicino T, Lanford R, Gerlich WH, Will H, Schaefer S. Conserved transactivating and pro-apoptotic functions of hepadnaviral X protein in ortho- and avihepadnaviruses. Oncogene. 2002;21:6606-6613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Song CZ, Bai ZL, Song CC, Wang QW. Aggregate formation of hepatitis B virus X protein affects cell cycle and apoptosis. World J Gastroenterol. 2003;9:1521-1524. [PubMed] |
| 42. | Su F, Theodosis CN, Schneider RJ. Role of NF-kappaB and myc proteins in apoptosis induced by hepatitis B virus HBx protein. J Virol. 2001;75:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 97] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 43. | Chami M, Ferrari D, Nicotera P, Paterlini-Bréchot P, Rizzuto R. Caspase-dependent alterations of Ca2+ signaling in the induction of apoptosis by hepatitis B virus X protein. J Biol Chem. 2003;278:31745-31755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 44. | Kim KH, Seong BL. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. EMBO J. 2003;22:2104-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 45. | Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071-22078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 133] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 46. | Chen HY, Tang NH, Li XJ, Zhang SJ, Chen ZX, Wang XZ. Transfection and expression of hepatitis B virus x gene and its effect on apoptosis in HL-7702 cells. World J Gastroenterol. 2004;10:959-964. [PubMed] |
| 47. | Tanaka Y, Kanai F, Kawakami T, Tateishi K, Ijichi H, Kawabe T, Arakawa Y, Kawakami T, Nishimura T, Shirakata Y. Interaction of the hepatitis B virus X protein (HBx) with heat shock protein 60 enhances HBx-mediated apoptosis. Biochem Biophys Res Commun. 2004;318:461-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 48. | Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Stürzbecher HW, Hoeijmakers JH, Harris CC. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012-6016. [PubMed] |
| 49. | Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707-14712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 253] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 50. | Gottlob K, Fulco M, Levrero M, Graessmann A. The hepatitis B virus HBx protein inhibits caspase 3 activity. J Biol Chem. 1998;273:33347-33353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 105] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 51. | Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, Woodgett JR, Penninger J, Richardson CD. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem. 2001;276:8328-8340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 52. | Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein inhibits transforming growth factor-beta -induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem. 2000;275:25858-25864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 159] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | Huo TI, Wang XW, Forgues M, Wu CG, Spillare EA, Giannini C, Brechot C, Harris CC. Hepatitis B virus X mutants derived from human hepatocellular carcinoma retain the ability to abrogate p53-induced apoptosis. Oncogene. 2001;20:3620-3628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 54. | Kim YC, Song KS, Yoon G, Nam MJ, Ryu WS. Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx-mediated apoptosis. Oncogene. 2001;20:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 55. | Lee YI, Kang-Park S, Do SI, Lee YI. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem. 2001;276:16969-16977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 56. | Li D, Chen X, Zhang W. The inhibition of apoptosis of hepatoma cells induced by HBx is mediated by up-regulation of survivin expression. J Huazhong Univ Sci Technolog Med Sci. 2003;23:383-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 57. | Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein activates a survival signaling by linking SRC to phosphatidylinositol 3-kinase. J Biol Chem. 2003;278:31807-31813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 58. | Kalra N, Kumar V. c-Fos is a mediator of the c-myc-induced apoptotic signaling in serum-deprived hepatoma cells via the p38 mitogen-activated protein kinase pathway. J Biol Chem. 2004;279:25313-25319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 59. | Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 448] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 60. | Slee EA, O'Connor DJ, Lu X. To die or not to die: how does p53 decide. Oncogene. 2004;23:2809-2818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 191] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 61. | Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230-2234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 481] [Article Influence: 15.5] [Reference Citation Analysis (1)] |
| 62. | Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish JA. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69:1851-1859. [PubMed] |
| 63. | Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 254] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 64. | Takada S, Kaneniwa N, Tsuchida N, Koike K. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene. 1997;15:1895-1901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 65. | Prost S, Ford JM, Taylor C, Doig J, Harrison DJ. Hepatitis B x protein inhibits p53-dependent DNA repair in primary mouse hepatocytes. J Biol Chem. 1998;273:33327-33332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 66. | Groisman IJ, Koshy R, Henkler F, Groopman JD, Alaoui-Jamali MA. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis. 1999;20:479-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 67. | Jia L, Wang XW, Harris CC. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 68. | Sohn S, Jaitovitch-Groisman I, Benlimame N, Galipeau J, Batist G, Alaoui-Jamali MA. Retroviral expression of the hepatitis B virus x gene promotes liver cell susceptibility to carcinogen-induced site specific mutagenesis. Mutat Res. 2000;460:17-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 69. | Jaitovich-Groisman I, Benlimame N, Slagle BL, Perez MH, Alpert L, Song DJ, Fotouhi-Ardakani N, Galipeau J, Alaoui-Jamali MA. Transcriptional regulation of the TFIIH transcription repair components XPB and XPD by the hepatitis B virus x protein in liver cells and transgenic liver tissue. J Biol Chem. 2001;276:14124-14132. [PubMed] |
| 70. | Takada S, Kaneniwa N, Tsuchida N, Koike K. Hepatitis B virus X gene expression is activated by X protein but repressed by p53 tumor suppressor gene product in the transient expression system. Virology. 1996;216:80-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 71. | Doitsh G, Shaul Y. HBV transcription repression in response to genotoxic stress is p53-dependent and abrogated by pX. Oncogene. 1999;18:7506-7513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 72. | Ogden SK, Lee KC, Barton MC. Hepatitis B viral transactivator HBx alleviates p53-mediated repression of alpha-fetoprotein gene expression. J Biol Chem. 2000;275:27806-27814. [PubMed] |
| 73. | Uchida T, Takahashi K, Tatsuno K, Dhingra U, Eliason JF. Inhibition of hepatitis-B-virus core promoter by p53: implications for carcinogenesis in hepatocytes. Int J Cancer. 1996;67:892-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 74. | Dandri M, Schirmacher P, Rogler CE. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246-5254. [PubMed] |
| 75. | Dandri M, Petersen J, Stockert RJ, Harris TM, Rogler CE. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J Virol. 1998;72:9359-9364. [PubMed] |
| 76. | Su Q, Schröder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 176] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 77. | Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci USA. 1994;91:674-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 245] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 78. | Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 993] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 79. | Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 990] [Cited by in RCA: 980] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 80. | Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1504] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 81. | Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 682] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 82. | Hall PA, Lane DP. Tumor suppressors: a developing role for p53. Curr Biol. 1997;7:R144-147. [RCA] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 83. | Puisieux A, Ji J, Guillot C, Legros Y, Soussi T, Isselbacher K, Ozturk M. p53-mediated cellular response to DNA damage in cells with replicative hepatitis B virus. Proc Natl Acad Sci USA. 1995;92:1342-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 84. | Lu X, Lane DP. Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes. Cell. 1993;75:765-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 574] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 85. | Poon RY, Jiang W, Toyoshima H, Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr-14/Tyr-15 phosphorylation after UV-induced DNA damage. J Biol Chem. 1996;271:13283-13291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 123] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 86. | Petrocelli T, Poon R, Drucker DJ, Slingerland JM, Rosen CF. UVB radiation induces p21Cip1/WAF1 and mediates G1 and S phase checkpoints. Oncogene. 1996;12:1387-1396. [PubMed] |
| 87. | May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621-7636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 443] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 88. | Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644-7655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 697] [Cited by in RCA: 752] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 89. | el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6088] [Cited by in RCA: 6305] [Article Influence: 197.0] [Reference Citation Analysis (0)] |
| 90. | Lu X, Burbidge SA, Griffin S, Smith HM. Discordance between accumulated p53 protein level and its transcriptional activity in response to u.v. radiation. Oncogene. 1996;13:413-418. [PubMed] |
| 91. | Albrecht JH, Meyer AH, Hu MY. Regulation of cyclin-dependent kinase inhibitor p21(WAF1/Cip1/Sdi1) gene expression in hepatic regeneration. Hepatology. 1997;25:557-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 92] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 92. | Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3502] [Cited by in RCA: 3561] [Article Influence: 107.9] [Reference Citation Analysis (0)] |
| 93. | Lowe SW, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 514] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 94. | Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1751] [Cited by in RCA: 1805] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 95. | Park US, Park SK, Lee YI, Park JG, Lee YI. Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1--> S transition via a p53-independent pathway in human hepatoma cells. Oncogene. 2000;19:3384-3394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 96. | Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 536] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 97. | Han J, Yoo HY, Choi BH, Rho HM. Selective transcriptional regulations in the human liver cell by hepatitis B viral X protein. Biochem Biophys Res Commun. 2000;272:525-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 98. | Livezey KW, Negorev D, Simon D. Hepatitis B virus-transfected Hep G2 cells demonstrate genetic alterations and de novo viral integration in cells replicating HBV. Mutat Res. 2000;452:163-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 99. | Kobayashi S, Matsushita K, Saigo K, Urashima T, Asano T, Hayashi H, Ochiai T. P21WAF1/CIP1 messenger RNA expression in hepatitis B, C virus-infected human hepatocellular carcinoma tissues. Cancer. 2001;91:2096-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 100. | Wu CG, Salvay DM, Forgues M, Valerie K, Farnsworth J, Markin RS, Wang XW. Distinctive gene expression profiles associated with Hepatitis B virus x protein. Oncogene. 2001;20:3674-3682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 101. | Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079-1085. [PubMed] |
| 102. | Akagi T, Ono H, Shimotohno K. Expression of cell-cycle regulatory genes in HTLV-I infected T-cell lines: possible involvement of Tax1 in the altered expression of cyclin D2, p18Ink4 and p21Waf1/Cip1/Sdi1. Oncogene. 1996;12:1645-1652. [PubMed] |
| 103. | Cereseto A, Diella F, Mulloy JC, Cara A, Michieli P, Grassmann R, Franchini G, Klotman ME. p53 functional impairment and high p21waf1/cip1 expression in human T-cell lymphotropic/leukemia virus type I-transformed T cells. Blood. 1996;88:1551-1560. [PubMed] |
| 104. | Mesnard JM, Devaux C. Multiple control levels of cell proliferation by human T-cell leukemia virus type 1 Tax protein. Virology. 1999;257:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 73] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 105. | Faktor O, Shaul Y. The identification of hepatitis B virus X gene responsive elements reveals functional similarity of X and HTLV-I tax. Oncogene. 1990;5:867-872. [PubMed] |
| 106. | Morozov A, Shiyanov P, Barr E, Leiden JM, Raychaudhuri P. Accumulation of human papillomavirus type 16 E7 protein bypasses G1 arrest induced by serum deprivation and by the cell cycle inhibitor p21. J Virol. 1997;71:3451-3457. [PubMed] |
| 107. | Ruesch MN, Laimins LA. Initiation of DNA synthesis by human papillomavirus E7 oncoproteins is resistant to p21-mediated inhibition of cyclin E-cdk2 activity. J Virol. 1997;71:5570-5578. [PubMed] |
| 108. | Butz K, Geisen C, Ullmann A, Zentgraf H, Hoppe-Seyler F. Uncoupling of p21WAF1/CIP1/SDI1 mRNA and protein expression upon genotoxic stress. Oncogene. 1998;17:781-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 109. | Fiedler N. Kombinierter Effekt des Hepatitis B Virus X Proteins und des Tumorsuppressorproteins p53 auf virale und zelluläre Promotoren [Diplomarbeit]. Gießen: Justus-Liebig-Universität 1996; . |
| 110. | Tarn C, Lee S, Hu Y, Ashendel C, Andrisani OM. Hepatitis B virus X protein differentially activates RAS-RAF-MAPK and JNK pathways in X-transforming versus non-transforming AML12 hepatocytes. J Biol Chem. 2001;276:34671-34680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |