Published online Jun 28, 2006. doi: 10.3748/wjg.v12.i24.3841
Revised: November 1, 2005
Accepted: November 10, 2005
Published online: June 28, 2006
AIM: To investigate whether Thy1 recognizes oval cells in the fetal liver and to characterize the cultured Thy1- selected cells from E14 rat livers.
METHODS: Thy1 populations were analyzed by fluorescence activated cell sorter analysis. Thy1 positive cells were isolated using magnetic beads. Hepatic markers were detected by Western blotting, immunocytochemistry and RT-PCR.
RESULTS: The percentage of Thy1-positive cells decreased during early development of fetal rat liver (E13-E16). E14 fetal livers contained 7.8% Thy1 positive cells, of which 61% were positive for α-fetoprotein (AFP) and 25% expressed albumin. The Thy1+ population expressed oval cell markers c-Kit and CXCR4, liver enriched-transcription factors HNF1α and HNF6, hepatocytic markers albumin, AFP and cytokeratin 18, and biliary marker cytokeratin 19. Thy1- selected cells formed only mesenchymal colonies when plated on collagen and in serum-containing media. Thy1 selected cells were able to form hepatic colonies positive for HNF1α, HNF6, albumin, AFP, cytokeratin 18, cytokeratin 19 and glycogen, when grown on STO feeder layers in serum free-media.
CONCLUSION: Oval cells positive for Thy1 are present in early liver embryonic stages.
- Citation: Isabel Z, Miri B, Einav H, Ella BL, Zamir H, Ran O. Isolation, characterization and culture of Thy1-positive cells from fetal rat livers. World J Gastroenterol 2006; 12(24): 3841-3847
- URL: https://www.wjgnet.com/1007-9327/full/v12/i24/3841.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i24.3841
Bipotential hepatic stem cells are found in murine and rat embryonic livers at stages E13 and E14, although at these embryonic stages a large majority of cells are already committed to the hepatocytic and bile duct lineage. Bipotential liver cells or hepatoblasts express an array of genes, such as α-fetoprotein (AFP), albumin, gamma-glutamyl transpeptidase (GGT), glutathione-S-transferase p and HNF6[1-3]. With maturation towards the hepatocytic lineage, fetal liver parenchymal cells have lower levels of AFP and higher levels of albumin, start expressing cytokeratin 18 (CK18) and break loose of GGT expression. Cells that differentiate towards biliary epithelial lineage retain GGT expression, express cytokeratins 7 and 19, c-kit and CD34 and stop expressing AFP and albumin[1-5]. Some liver-specific genes, such as glutamine synthetase, tyrosine aminotransferase (TAT), phosphoenolpyruvate carboxykinase and α2-microglobulin appear later in hepatocytes, at birth or even after birth. Recently, several groups have isolated bipotential fetal progenitor cells from murine and rat origin, which form colonies of both hepatocytes and bile duct cells, identified by albumin and cytokeratin 19 (CK19) expression, respectively[6,7].
There is controversy over the identity of adult liver stem cells, and whether they exist in the adult liver. One of the candidates is the oval cells. However, oval cells are only detected after certain protocols, such as carcinogenic insults to the liver, followed by partial hepatectomy[8]. There are suggestions that oval cells are bone marrow cells that migrate to the liver and become hepatocytes. Reconstitution of sublethally-irradiated male mice with female bone marrow, followed by a protocol that results in oval cell proliferation, showed that oval cell-derived hepatocytes are of donor origin[9]. Moreover, the homing molecule for hematopoietic stem cells to the bone marrow, stroma-derived factor 1 (SDF-1), is induced on hepatocyte surface in livers treated with oval cell proliferation-inducing protocols, and the oval cells express the CXCR4, the receptor for SDF-1[10]. If indeed oval cells are of bone marrow origin, it may explain the expression of common markers in both oval cells and hematopoietic cells, such as Thy1, CD34 and c-kit. Several current studies have shown that bone marrow cells that arrive at the liver do not differentiate into hepatocytes, but undergo fusion with hepatocytes[11].
Recently, one group has investigated the presence of oval cells in the fetal liver, since expression of Thy1 is never determined in fetal liver hepatoblasts[12,13]. This group isolated Thy1 positive cells from E16-E20 livers, and stained these cells with hepatocytic marker CK18[12]. They found that 0.5%-1% of the cells are positive for both Thy1 and CK18 and speculated that these double-positive cells may be hepatic progenitor cells[12]. Other markers for oval cells are c-kit, oncostatin M receptor β and CXCR4 and dlk[10,14,15]. Dlk is a marker for both murine hepatoblsts and murine oval cells, and its expression is the strongest at E10.5 and down regulated after E16.5[16].
The purpose of the present study was to determine whether oval cells are present in early embryonic fetal liver, and to characterize the cultured Thy1- positive cells from E14 fetal livers.
Fischer 344 rats with known durations of pregnancy were maintained in the animal facility of the Tel Aviv Sourasky Medical Center on a standard rat chow diet in a 12 h light and dark cycle. By convention, the first day of gestation was defined as d 0. Use of animals was in accordance with the NIH policy on the care and use of laboratory animals and approved by the Animal Care and Use Committee of the Tel Aviv Sourasky Medical Center.
Cell suspensions enriched for liver parenchymal cells were isolated as described previously with minor modifications[17]. Livers from E13, E14 and E16 embryos were removed and placed into ice cold, Ca2+-free Hank’s balanced salt solution (HBSS) containing 0.8 mmol/L MgCl2 and 20 mmol/L HEPES (Sigma Chemical Company, Israel), pH 7.3 (modHBSS). After all tissues were collected and non-hepatic tissue was removed, HBSS-5 mol/L EGTA was added to a final EGTA concentration of 1 mmol/L. The livers were gently triturated 6-8 times to partially disaggregate the tissue and then centrifuged at 400 r/min for 5 min at 4°C (all subsequent centrifugation steps were done at the same settings). The pellet of cells and tissue were resuspended in 35 mL modHBSS containing 0.08 g collagenase type 1 (Worthington Biochemical Corporation, NJ, USA) and 1 mmol/L MgCl2, gently triturated and stirred at 37°C for 15 min in an Erlenmeyer flask. The dispersed cells were pooled, centrifuged and resuspended in modHBSS. Cell number and viability were determined by hemacytometer and trypan blue exclusion.
The freshly isolated liver cells from fetal livers were stained for 1 h on ice with Thy1-fluorescein isothiocyonate (FITC) (Serotec, Oxford, United Kingdom) diluted in DMEM + 100 mL/L fetal calf serum (FCS) (Biological Industries, Bet Haemek, Israel). At the end of the incubation, the cells were washed and fixed in 5 g/L paraformaldehyde in phosphate buffered saline (PBS). Thy1-positive cells were analyzed using a Becton-Dickson fluorescence activated cell sorter (FACS) and the results were analyzed using the WinMDI software. For double staining of Thy1 and α-fetoprotein, the cells were stained with Thy1-phycoerythrin (PE)(1:100), fixed with solution A of Intrastain kit (Dako, Glostrup, Denmark), permeabilized with solution B, and stained for intracellular expression with antibodies to rat α-fetoprotein-FITC (1:80) (Nordic, Tilburg, The Netherlands). Thy1 and α-fetoprotein were analyzed using a Becton-Dickson FACS and the results were analyzed using CellQuest software. For double labeling of cell surface with dlk and oncostatin receptor β, and intracellular staining of AFP, we used antibodies to dlk (Santa Cruz Biotechnologies, Santa Cruz, CA) and to mouse OSMRβ (R&D Systems, Minneapolis, MN), followed by anti-goat-PE conjugated antibody (Jackson Laboratories, Bar Harbor, ME) and staining with AFP-FITC.
The freshly isolated fetal liver cells were selected using an EasySep kit from StemCell Technologies, Vancouver, Canada. Briefly, cells were incubated for 15 min at room temperature with 3 mg/L Thy1-FITC (Serotec, Oxford, United Kingdom) in PBS containing 20 mL/L FCS and 1 mmol/L EDTA. Cells were washed once with excess buffer, then incubated with EasySep FITC selection cocktail for another 15 min, and then with the EasySep magnetic nanoparticles for 10 min. Buffer was added to cells at total volume of 2.5 mL, and the labeled cells were placed for 5 min inside the Violet Magnet (StemCell Technologies, UK). Unlabeled cells were poured off from the tube. The procedure was repeated two more times. The selected cells were resuspended in buffer and counted, and used for further characterization or for plating.
Thy1-selected cells were plated at two densities per well in 6- well plates (approximately 1000 and 2000 cells/cm2). The plates were coated with collagen type I. The media used to culture the cells were as follows. Medium 1: DMEM supplemented with 100 mL/L FCS (Biological Industries, Bet Haemek, Israel), 50 μg/L EGF (PeproTech Asia/CytoLab, Rehovot, Israel) and 10 mg/L insulin (Sigma, St. Louis, MI)[18]; medium 2: DMEM supplemented with 10 μg/L oncostatin M and 10 μg/L flt-3 ligand (PeproTech Asia/CytoLab, Rehovot, Israel)[19]; medium 3: DMEM supplemented with 100 mL/L FCS, 1mg/L insulin, 10-7 mol/L dexamethasone, 10 mmol/L nicotinamide, 50 μmole/L β-mercaptoethanol, 50 μg/L HGF (PeproTech Asia/CytoLab, Rehovot, Israel), 20 μg/L EGF[6]; medium 4: DMEM supplemented with 100 mL/L FCS.
The cells were allowed to grow for 2 wk with the medium changed every three days. The colonies were stained with 2.5 g/L crystal violet solution and counted. The experiments were done at least three times.
STO cells were a kind gift of Professor Lola Reid (UNC, Chapel Hill, NC). The cells were cultured in DMEM ( F12 media 1: 1) supplemented with 100 mL/L FCS, 10 mL/L dimethylsulfoxide (DMSO), 100 mg/L penicillin and 100 mg/L streptomycin.
The STO cells were growth arrested for 3 h with 10 mg/L mitomycin C (Sigma, St. Louis, MI), then plated on 1 g/L gelatin-coated plates at a density of 60 × 103 cells/cm2 in the medium described above and cultured for 2 d prior to seeding Thy1- selected cells on them.
Thy1-selected cells from e14 fetal livers were plated on STO feeder layers at a density of 4000 cells/cm2 and cultured for 2 wk in hormonally defined medium (HDM)[20] consisting of DMEM supplemented with 100 mg/L penicillin/streptomycin, 2 g/L bovine serum albumin, 610 mg/L nicotinamide, 740 μg/L ZnSO4× 7H2O, 20 μg/L CuSO4× 5H2O, 5 mmol/L glutamine, 5 mg/L insulin, 5 mg/L iron-saturated transferrin, 5 μg/L selenous acid and 10-7 mole/L dexamethasone. All the medium additives were obtained from Sigma, St. Louis, MI.
Total protein was extracted by incubation for 30 min on ice in lysis buffer (250 mmol/L sucrose, 5 mmol/L MgCl2, 10 mmol/L Tris pH 8.0, 5 g/L Triton X-100 and 1 mmol/L PMSF), and insoluble material pelleted at 17 000 g. The extracts were normalized to total protein content, determined using Bradford reagent (Sigma, St. Louis, MI) and boiled for 5 min in sample buffer containing SDS and β-mercaptoethanol. Protein (10-20 μg per lane) was separated by SDS-PAGE (BioRad Protean II minigel) in 100 g/L acrylamide gels and blotted onto Hybond C extra (Amersham, Arlington Heights, IL) for 1 h at 250 mA constant current. The blots were blocked overnight in Tris-saline buffer pH 7.5, containing 0.5 g/L Tween-20 and 50 g/L milk, and incubated with antibodies to rat albumin (Sigma, St. Louis, MI) and AFP (Santa Cruz Laboratories, Santa Cruz, CA), washed and incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibody (Jackson Laboratories, Bar Harbor, ME). The blots were then subjected to chemiluminescent detection and flurorography using X-ray film (NEN Life Science Products, Boston, MA).
Cells were fixed with 95 mL/L ethanol for 10 min at 4°C, and incubated with a blocking solution containing 100 mL/L FCS for 30 min at room temperature. Cells were then incubated with primary antibodies to rat albumin (1:400) (Sigma, St. Louis, MI), mouse AFP (1:200), CK19 (Novacastra) for 1 h at room temperature, washed three times, followed by addition of anti-rabbit second antibody conjugated to Cy3 (1:100) (Jackson Laboratories, Bar Harbor, ME) for another hour at room temperature. Cells were washed three times and stained cells were visualized under Axiovert 25 microscope (Zeiss, Germany) with fluorescent attachments.
Glycogen granules in the cultured cells were detected using a periodic acid-Schiff (PAS) staining kit according to the manufacturer’s instructions (Sigma, St. Louis, MI).
Total RNA was extracted from freshly isolated E14 and Thy1+ selected cells using EZ RNA kit (Biological Industries, Bet Haemek, Israel), treated with DNAse and cDNA was reverse-transcribed. For reverse transcription, 1-2 μg of total RNA was used. The cDNA obtained was used for PCR, using the following oligonucleotide primers[21]: HNF-1α: the 5’ primer is 5’-AGCTGCTCCTCCATCATCAGA -3’, the 3’ primer is 5’-TGTTCCAAGCATTAAGTTTTCTATTCTAA -3’; HNF4α: the 5’ primer is 5’-CTTCCTTCTTCA-TGCCAG -3’, the 3’ primer is 5’-ACACGTCCCCAT-CTGAAG -3’; HNF6: the 5’ primer is 5’-AACTCCCAGCAAG-GACTTCCCCACT-3’, the 3’ primer is 5’-CCATCTGCCCTGAAT-TACTTCCATTGC-3’; cytokeratin 18 (CK18): the 5’ primer is 5’-GCCCTGGACTCCAGCAACT -3’ and the 3’ primer is 5’-ACTTTGCCATCCACGACCTT -3’; cytokeratin 19 (CK19): the 5’ primer is 5’-ACCATGCAGAACCTGAACGAT- 3’ and the 3’ primer is 5’- CACCTCCAGCTCGCCATTAG-3’; albumin: the 5’ primer is 5’-CTGGGAGTGTGCA-GATATCAGAGT- 3’ and the 3’ primer is 5’-GAGAAG-GTCACCAAGTGCTGTAGT-3’; alpha-fetoprotein: the 5’ primer is 5’-GTCCTTTCTTCCTCCTGGAGAT- 3’ and the 3’ primer is 5’-CTGTCACTGCTGATTTCTCTGG-3’; oncostatin M receptor: the 5’ primer is 5’-CCCCTGTGAG-GCCGAGGACCGGC-3’ and the 3’ primer is 5’-TGGCTTTACAATTGATGTTTGTCCCTG-3’; stroma-derived factor 1(SDF1): the 5’ primer is 5’–ATTCTTTGAGAGCCATGTCGC-3’ and the 3’ primer is 5’-CCTTGAGCTGAGTGACTCTCG-3’; C-kit: the 5’ primer is 5’-AGCAAGAGTTAAC-GATTCCGGAG-3’ and the 3’ primer is 5’-CCAGAAAGGTGTAAGTGCCTCCT-3’; CXCR4: the 5’ primer is 5’-CCCTCGAGATGGAA-ATATACAC-3’ and the 3’ primer is 5’-GCTCTAGATTAGC-TGGAGTGAAA-3’. For glyceraldehydehyde-3-phosphate-dehydrogenase (GAPDH), the 5’ primer is 5’-ACCACAGTCCATGCCATCAC-3’ and the 3‘ primer is 5’-TCCACCACCCTGTTGCTGTA-3’. For PCR, 1.5 μL of the reverse transcription reaction was used, and the reaction mixture also contained 0.4 μmol/L of each primer, 200 μmol/L dNTP, 10 mmol/L Tris-HCl (pH = 8.3), 50 mmol/L KCl, 1.1 mmol/L MgCl2, 0.1 g/L gelatin and 2.5 U RED Taq DNA polymerase (Sigma, St. Louis, MI). The mixture was denatured for 1 min at 94°C. The PCR was started at 94°C for 10 s for the denaturation step, at 53°C for 30 s for the annealing step and at 72°C for 1 min for the elongation step. Thirty-five cycles were used to amplify the hepatic markers. The PCR products were finally separated in a 20 g/L agarose gel containing ethidium bromide.
We assessed the expression of oval cell markers, Thy1 and oncostatin receptor M β (OSMRβ), and fetal hepatoblast marker dlk in E13, E14 and E16 fetal livers (Table 1). The expression of all three markers decreased with liver development (Table 1). The largest percentage of Thy1 positive cells was in E13 rat livers (10%), and the percentage of Thy1-positive cells decreased with liver development, so that there were 7.8% Thy1 positive cells in E14 livers, and only 2.3% Thy1 positive cells in E16 livers (Table 1). We did not continue to test the percentages of Thy1 positive cells in later stages, since the peak of hematopoiesis in fetal rat liver was at E17 and E18, and Thy1- positive cells at those stages might be hematopoietic cells rather than hepatoblasts.
| Embryonal stage | Thy1 positive cells (%) | Dlk positive cells (%) | OSM receptor β positive cells (%) |
| E13 | 10.5 ± 2.0 | 10.8 | 5.3 |
| E14 | 7.8 ± 0.5 | 5.3 ± 1.1 | 3.6 ± 1.5 |
| E16 | 2.3 ± 0.6 | Not done | Not done |
We defined the positive populations for both Thy1 and α-fetoprotein in E14 fetal livers. In the E14 total liver population, 4.8% of the cells were positive for both Thy1 and α-fetoprotein, while 3 % were positive for Thy1 only (Table 1). There 2.7% of cells positive for dlk and AFP and 1.6% of cells positive for OSMRβ and AFP in the E14 livers.
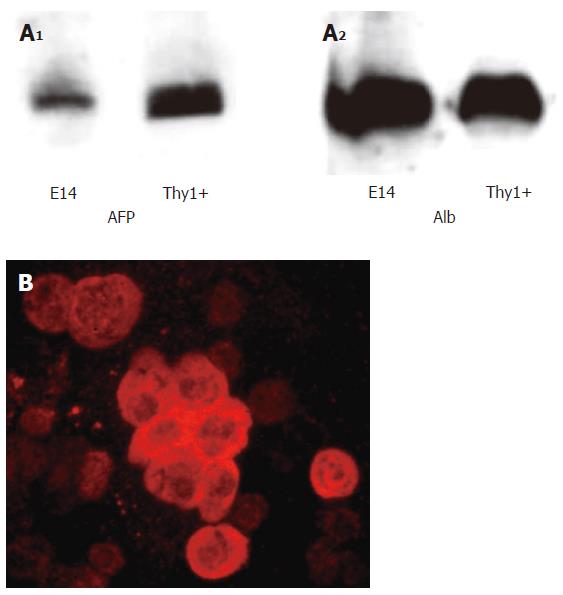
We obtained 11 × 106 single cell suspension cells from per E14 liver, with a viability > 95% determined by trypan blue exclusion. We selected Thy1- positive cells from the single cell suspensions of E14 liver cells using nanoparticles and magnet selection of Thy1-FITC labeled cells. We determined AFP and albumin expression by Western blots using freshly isolated E14 liver cells and Thy1- selected cells. Thy1-positive cells expressed slightly more AFP than E14 cells, while albumin expression was similar in the two cell populations (Figure 1A). We next determined the percentages of albumin positive cells in the Thy1- selected cells from E14 and E16 livers, using immunocytochemistry staining on cytospun cells. Albumin expression in E14 Thy1- selected cells is shown in Figure 1B. We counted albumin positive cells in 20 fields and found that 25% ±8% of Thy1+ from E14 livers were albumin positive and 18% ± 9% of Thy1+ cells from E16 livers expressed albumin.
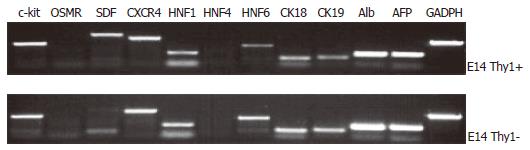
We also determined the expression of oval cell markers c-kit, OSMRβ and CXCR4, liver- enriched transcription factors HNF1α, HNF4α and HNF6, hepatocytic markers AFP, albumin, CK18, and biliary epithelial marker CK19 in Thy1+ and Thy1- cells from E14 livers. We observed that both cell populations were positive for c-kit, CXCR4, HNF1 and HNF6, albumin, AFP, CK18 and CK19, while only the Thy1+ cells were positive for SDF1α, and we could not detect OSMRβ and HNF4 in either cell population (Figure 2).
We plated the Thy1-selected cells at clonal densities on collagen type I-coated dishes and in four different media, as described in Materials and Methods. After two weeks, colonies were stained with crystal violet solution and counted. Both medium 2 and medium 3 were beneficial for development of colonies (Table 2). However, the morphology of the cells was not typical of hepatoblasts, but rather of fibroblasts (data not shown). We isolated four clones from two separate experiments of cells grown in medium 3 and on collagen I, and tested them by RT-PCR for hepatic markers. The only hepatic marker detected was CK18 (data not shown).
| Media | Colonies (n) | Colonies (n) |
| Medium 1 | 7.3 ± 2 | ND |
| Medium 2 | 11 ± 6.8 | ND |
| Medium 3 | 9.5 ± 2 | 28 ± 8.7 |
| Medium 4 | ND | 54 ± 1.8 |
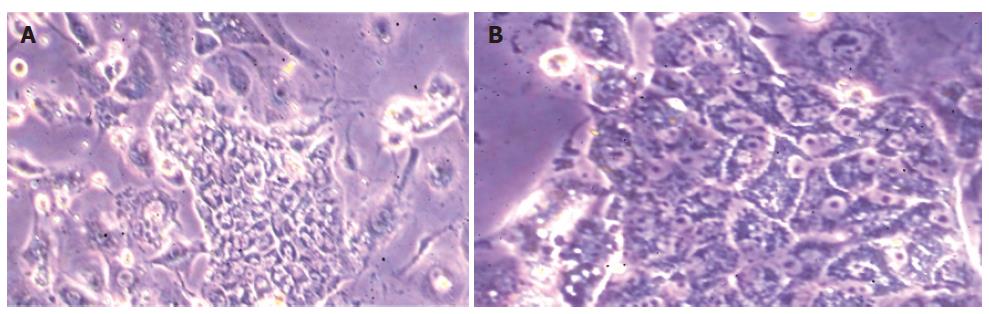
Since 25% of the Thy1- selected cells expressed albumin, we thought that in the above conditions, the albumin-positive cells probably did not survive. Therefore, we plated Thy1-selected cells on collagen type I-coated plates and in DMEM with 100 mL/L FCS, at a density of 500 000 cells/well in 6-well plates. After one and two days in culture, typical fetal hepatoblast colonies were observed and stained positive for albumin (data not shown). However, the colonies did not survive for longer periods, but peeled off the plates. Since hepatic fetal cells could grow on feeder layers[7], we plated Thy1- selected cells on STO feeders and in serum free-medium. The cells survived very well under these conditions for two weeks, and formed typical fetal hepatoblast colonies (Figure 3). These colonies were positive for albumin, AFP, CK18 and glycogen (Figure 4). We determined that 0.6% ± 0.17% of the total Thy1 positively-plated cells formed albumin-positive colonies. However, there were numerous other colonies on the plates, with fibroblast morphology similar to the colonies obtained when the cells were cultured on collagen I and in media 1-4 (data not shown).
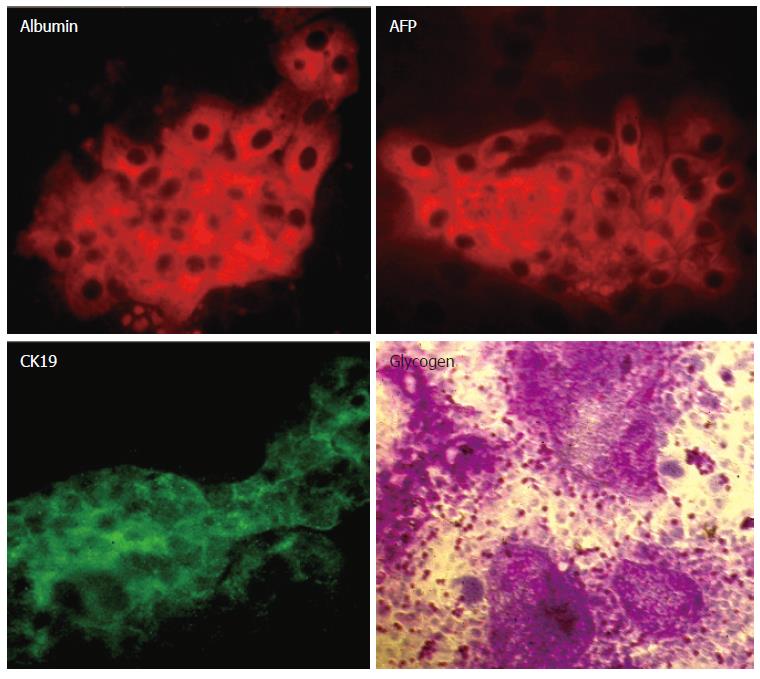
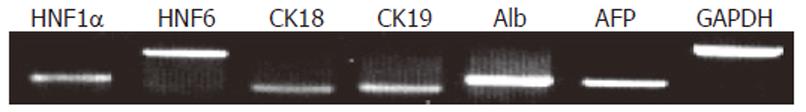
We examined the expression of mRNA for hepatocytic markers in two weeks- old cultures of Thy1-selected cells grown on STO feeders. We found that there was expression of liver enriched-transcription factors HNF1α and HNF6, as well as hepatic markers albumin, AFP, CK18 and biliary marker CK19 (Figure 5). The expression of OSMRβ was very weak in the colonies derived from Thy1 positive cells, while transcription factor HNF4α was not detected (data not shown).
In the present study, early rat fetal livers contained Thy1-positive cells, and the percentage of Thy1- positive cells decreased between E13 and E16. E14 fetal livers contained about 8% Thy1-positive cells, of which 61% were positive for AFP and 25% were positive for albumin. The Thy1- positive cells expressed oval cell markers c-kit and CXCR4, liver enriched-transcription factors HNF1α and HNF6, hepatocytic markers albumin, α fetoprotein and CK18, and biliary marker CK19. We were able to culture Thy1- selected cells from E14 livers for two weeks on STO feeder layers, and observed several types of colonies, both mesenchymal and hepatic colonies.
In the last few years, hepatic stem cells from various sources, both fetal and adult livers, have been studied extensively. Methods used to isolate the cells from murine and rat fetal livers include FACS sorting using several positive and negative markers[6,7]. When the isolated cells are cultured, they form colonies that contain both hepatocytes and bile duct epithelial cells.
We hypothesized that in addition to the fetal hepatoblasts, which could differentiate into hepatocytes and bile duct epithelial cells during fetal liver development, fetal liver might contain additional stem cells, the “oval” cells. We may surmise that cells positive for oval cell markers Thy1 and AFP might be more numerous in the early stages of fetal liver development and decrease in later stages, so that the oval cell numbers in the adult livers are very few, and in fact difficult to detect unless oval cells proliferate due to carcinogenic injuries.
Thy1 has been shown to be present on rat oval cells, one kind of the putative liver stem cells in the adult liver[9]. Thy1 (CD90) is a cell surface molecule, present on hematopoietic stem cells, subsets of T cells, bone marrow cells, fibroblasts and cells in the brain[22,23]. Thy1 may be a receptor for an unknown ligand, and may play a role in cellular recognition and adhesion [20,21].
Adult rat liver oval cells, in addition to Thy1, expressing AFP, cytokeratin 19 and GGT, are negative for hepatic stellate cell marker, desmin[9]. Other markers for oval cells are c-kit, oncostatin M receptor β and CXCR4[10,14]. Recent studies have shown that Thy1 is present not only on adult rat oval cells, but also in rat E16 fetal liver, and a small percent of Thy1 positive cells also express hepatocytic marker CK18[12], suggesting that the Thy1 + CK18 + are hepatic stem cells. The same group isolated Thy1 positive cells from E16, E18, E20 and E22, and found that the cells are also positive for albumin, AFP and CK18[13]. Long-term cultures (one month) of human fetal hepatocytes also express Thy1, as well as other oval cell and hematopoietic markers such as CD34 and OV-6. In addition, these cells assume a blast-like morphology and express hepatoblast markers AFP, CK19 and CK18[24].
Rat fetal liver rudiment appears at E10, E13 and E14, there are still numerous bipotential hepatic stem cells, though there are also cells committed towards the hepatocyte and biliary epithelial cell lineage[25]. Since the rat fetal liver is both a hepatic and a hematopoietic organ, we tested the expression of Thy1, a hematopoietic marker, before the peak of hematopoiesis in fetal liver, which occurred at E17 and lasted until birth. We found that there were Thy1 positive cells in E13 livers, and the percentage of Thy1 positive cells decreased between E13 and E16.
We chose E14 fetal livers as the ideal stage for isolating oval cells, since the livers contain a large number of cells, and there are many Thy1+ AFP+ cells. Moreover, we observed that Thy1 cells selected from E14 livers expressed numerous hepatic and biliary epithelial markers and liver-enriched transcription factors, as well as oval cell markers c-kit and CXCR4, and OSMRβ detected by FACS analysis, but not by RT-PCR[10]. However, we did detect OSMRβ in colonies of Thy1 positive cells, suggesting that its expression is very weak in the original cells. The Thy1-selected cells were not homogenous and contained both oval cells (Thy1+ AFP+) and mesenchymal cells, (Thy1+ AFP-). When we cultured the cells on collagen I and in various media containing serum, we observed only mesenchymal cell colonies and some epithelial colonies that were positive for CK18. However, when we cultured the Thy1 positive cells on STO feeder layers, we detected formation of hepatic colonies expressing albumin and AFP. The original Thy1+ cells and colonies derived from Thy positive cells were negative for HNF4. It was reported that oval cells are negative for HNF4, and only start expressing this transcription factor when they differentiate into hepatocytes[26], suggesting that fetal oval cells may not undergo complete differentiation into hepatocytes under our culture conditions.
In mice, contrary to rat and human livers, Thy1 appears to identify only mesenchymal cells both in adult and in fetal liver[27,28]. Thy1 positive cells isolated from murine fetal livers have been shown to support differentiation of CD49f positive cells to hepatocytes[28]. The Thy1 positive cells comprise three different populations, one positive for α-smooth muscle actin, another positive for desmin, both markers of hepatic stellate cells, and a third population negative for both[28]. The murine Thy1 positive cells are negative for AFP, cytokeratin 19 and albumin[28].
In conclusion, Thy1 identifies putative oval cells during early fetal liver development, but is also present on other cells such as mesenchymal cells.
S- Editor Pan BR L- Editor Wang XL E- Editor Bai SH
| 1. | Shiojiri N, Lemire JM, Fausto N. Cell lineages and oval cell progenitors in rat liver development. Cancer Res. 1991;51:2611-2620. [PubMed] |
| 2. | Fausto N, Lemire JM, Shiojiri N. Cell lineages in hepatic development and the identification of progenitor cells in normal and injured liver. Proc Soc Exp Biol Med. 1993;204:237-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 52] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 3. | Germain L, Blouin MJ, Marceau N. Biliary epithelial and hepatocytic cell lineage relationships in embryonic rat liver as determined by the differential expression of cytokeratins, alpha-fetoprotein, albumin, and cell surface-exposed components. Cancer Res. 1988;48:4909-4918. [PubMed] |
| 4. | Crosby HA, Kelly DA, Strain AJ. Human hepatic stem-like cells isolated using c-kit or CD34 can differentiate into biliary epithelium. Gastroenterology. 2001;120:534-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 5. | Fujio K, Evarts RP, Hu Z, Marsden ER, Thorgeirsson SS. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab Invest. 1994;70:511-516. [PubMed] |
| 6. | Suzuki A, Zheng Y, Kondo R, Kusakabe M, Takada Y, Fukao K, Nakauchi H, Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 212] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 7. | Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc Natl Acad Sci U S A. 2000;97:12132-12137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 178] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Paku S, Schnur J, Nagy P, Thorgeirsson SS. Origin and structural evolution of the early proliferating oval cells in rat liver. Am J Pathol. 2001;158:1313-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 206] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 9. | Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, Murase N, Boggs SS, Greenberger JS, Goff JP. Bone marrow as a potential source of hepatic oval cells. Science. 1999;284:1168-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1795] [Cited by in RCA: 1669] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 10. | Hatch HM, Zheng D, Jorgensen ML, Petersen BE. SDF-1alpha/CXCR4: a mechanism for hepatic oval cell activation and bone marrow stem cell recruitment to the injured liver of rats. Cloning Stem Cells. 2002;4:339-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 175] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Olson S, Grompe M. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1114] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 12. | Fiegel HC, Park JJ, Lioznov MV, Martin A, Jaeschke-Melli S, Kaufmann PM, Fehse B, Zander AR, Kluth D. Characterization of cell types during rat liver development. Hepatology. 2003;37:148-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Fiegel HC, Kluth J, Lioznov MV, Holzhüter S, Fehse B, Zander AR, Kluth D. Hepatic lineages isolated from developing rat liver show different ways of maturation. Biochem Biophys Res Commun. 2003;305:46-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Okaya A, Kitanaka J, Kitanaka N, Satake M, Kim Y, Terada K, Sugiyama T, Takemura M, Fujimoto J, Terada N. Oncostatin M inhibits proliferation of rat oval cells, OC15-5, inducing differentiation into hepatocytes. Am J Pathol. 2005;166:709-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Tanimizu N, Nishikawa M, Saito H, Tsujimura T, Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J Cell Sci. 2003;116:1775-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 272] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 16. | Tanimizu N, Tsujimura T, Takahide K, Kodama T, Nakamura K, Miyajima A. Expression of Dlk/Pref-1 defines a subpopulation in the oval cell compartment of rat liver. Gene Expr Patterns. 2004;5:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Brill S, Zvibel I, Reid LM. Expansion conditions for early hepatic progenitor cells from embryonal and neonatal rat livers. Dig Dis Sci. 1999;44:364-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Strick-Marchand H, Weiss MC. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology. 2002;36:794-804. [PubMed] |
| 19. | Yang L, Li S, Hatch H, Ahrens K, Cornelius JG, Petersen BE, Peck AB. In vitro trans-differentiation of adult hepatic stem cells into pancreatic endocrine hormone-producing cells. Proc Natl Acad Sci U S A. 2002;99:8078-8083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 363] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 20. | Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133-1149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 366] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 21. | Schwartz RE, Reyes M, Koodie L, Jiang Y, Blackstad M, Lund T, Lenvik T, Johnson S, Hu WS, Verfaillie CM. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J Clin Invest. 2002;109:1291-1302. [PubMed] |
| 22. | Walsh FS, Ritter MA. Surface antigen differentiation during human myogenesis in culture. Nature. 1981;289:60-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | He HT, Naquet P, Caillol D, Pierres M. Thy-1 supports adhesion of mouse thymocytes to thymic epithelial cells through a Ca2(+)-independent mechanism. J Exp Med. 1991;173:515-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Lázaro CA, Croager EJ, Mitchell C, Campbell JS, Yu C, Foraker J, Rhim JA, Yeoh GC, Fausto N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 25. | Brill S, Holst P, Sigal S, Zvibel I, Fiorino A, Ochs A, Somasundaran U, Reid LM. Hepatic progenitor populations in embryonic, neonatal, and adult liver. Proc Soc Exp Biol Med. 1993;204:261-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Nagy P, Bisgaard HC, Thorgeirsson SS. Expression of hepatic transcription factors during liver development and oval cell differentiation. J Cell Biol. 1994;126:223-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 180] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Fujikawa T, Hirose T, Fujii H, Oe S, Yasuchika K, Azuma H, Yamaoka Y. Purification of adult hepatic progenitor cells using green fluorescent protein (GFP)-transgenic mice and fluorescence-activated cell sorting. J Hepatol. 2003;39:162-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Hoppo T, Fujii H, Hirose T, Yasuchika K, Azuma H, Baba S, Naito M, Machimoto T, Ikai I. Thy1-positive mesenchymal cells promote the maturation of CD49f-positive hepatic progenitor cells in the mouse fetal liver. Hepatology. 2004;39:1362-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |