Published online Apr 14, 2006. doi: 10.3748/wjg.v12.i14.2250
Revised: October 10, 2005
Accepted: November 18, 2005
Published online: April 14, 2006
AIM: To investigate whether krÜppel-like factor 6 (KLF6) plays an important role in the development and/or progression of colorectal cancer.
METHODS: A total of 123 formalin-fixed and paraffin-embedded colorectal cancer specimens were analyzed by immunohistochemistry using tissue microarray for the expression of KLF6 protein. The specimens were collected over a 3-year period in the laboratories at our large teaching hospital in Seoul, Republic of Korea. The correlation of KLF6 expression with clinicopathologic parameters was analyzed by χ2 test and Bartholomew test.
RESULTS: Normal colonic epithelium showed weak to moderate expression of KLF6, whereas reduced KLF 6 expression or loss of KLF6 expression was seen in 45 (36.6%) of the 123 colorectal carcinoma specimens. Interestingly, aberrant expression of KLF6 was detected in 25 (43.1%) of 58 cases with metastasis to regional lymph node and in 31 (47.0%) of 66 tumors more than 5 cm in size. Statistically, loss of KLF6 expression was significantly associated with tumor size (P < 0.05). However, there was no significant correlation between KLF6 expression and Dukes’ stage (Bartholomew test, P > 0.05), tumor location and lymph node metastasis (χ2 test, P > 0.05).
CONCLUSION: Loss of KLF6 expression may be a common and early event in colorectal carcinogenesis.
- Citation: Cho YG, Choi BJ, Song JW, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Aberrant expression of krÜppel-like factor 6 protein in colorectal cancers. World J Gastroenterol 2006; 12(14): 2250-2253
- URL: https://www.wjgnet.com/1007-9327/full/v12/i14/2250.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i14.2250
Colorectal cancer is the second leading cause of cancer-related death in the Western world[1]. In Korea, it accounts for an estimated 11.2% of all malignancies, with 11.6% in the male population and 10.7% in the female population[2]. Thus, colorectal cancer remains a significant contributor to the world’s health burden. At a molecular level, great progress has been made in the last two decades in identification and characterization of the genetic changes involved in the malignant colorectal transformation process. Thus, the concept of multistage carcinogenesis is now widely accepted as a consequence of multiple genetic alterations accumulated in cancer cells [3]. However, the molecular mechanisms underlying dysregulated cell growth in colorectal cancer remain the subject of intensive investigation.
Krüppel-like factor 6 (KLF6) is a ubiquitously expressed zinc finger transcription factor that is part of a growing KLF family. The KLF family is broadly involved in growth-related signal transduction, cell proliferation, development and apoptosis, as well as angiogenesis[4,5]. In functional analysis, wild-type KLF6 up-regulates the cell cycle inhibitor p21 in a TP53-independent manner and suppresses growth, whereas tumor-derived KLF6 mutants fail to upregulate p21 or suppress proliferation in prostatic and non-small cell lung cancer cells[6,7]. In addition, introduction of KLF6 disrupts cyclin D1-dependent kinase 4 complexes and forces the redistribution of p21, which promotes G1 cell cycle arrest[8]. Furthermore, KLF6 plays a role as an inhibitor of cell proliferation by counteracting the function of the c-Jun protooncoprotein[9].
Recently, genetic alterations of KLF6 gene have been identified in several human cancers, including colorectal cancer, hepatocellular carcinoma, prostate and gastric cancer[10-13]. Reduced expression of KLF6 protein has been found in non-small cell lung cancers and glioblastomas[7,14]. Interestingly, the survey of gene expression in a panel of 60 NCI cancer cell lines showed that expression of KLF6 mRNA was reduced in colon cancer cell lines[15]. However, there is no report describing the expression pattern of KLF6 protein in colorectal cancers.
In the present study, to determine whether altered expression of the KLF6 protein is involved in colorectal cancer development, we analyzed the expression pattern of KLF6 on a series of 123 colorectal cancer specimens.
One hundred and twenty-three formalin-fixed and paraffin-embedded colorectal cancer specimens collected between 2002 and 2003 were enrolled in this study. No patient had a family history of colorectal cancer. Tumor stage was classified according to Dukes’ criteria. Thirteen patients were classified as Dukes’ A, 47 as Dukes’ B, 55 as Dukes’ C and 8 as Dukes’ D[16]. Two pathologists screened histological sections and selected areas of the representative tumor cells. Three tissue cores (0.6 mm in diameter) were taken from each tumor sample and placed in a new recipient paraffin block using a commercially available microarray instrument (Beecher Instruments, Micro-Array Technologies, Silver Spring, MD) as previously described[17]. One cylinder of normal colonic mucosa adjacent to each tumor was also transferred to the recipient block.
The primary polyclonal rabbit anti-KLF6 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used. Immunostaining was performed on microarray tissue sections with a tyramide signal amplification kit (NEN Life Science, Boston, MA) for signal intensification. Antigen retrieval was performed by microwave heating in a citrate buffer (pH 6.0). Other procedures were performed as previously described[18]. Reaction products were developed with diaminobenzidine (Sigma, St Louis, MO) and counterstained with hematoxylin. Because all the corresponding normal colonic mucosae showed KLF6 expression, tumors were considered as negative when nuclear and cytoplasmic staining was seen in less than 20% of cancer cells. We calculated the KLF6 positive and negative cancer cells in three representative areas at X200 magnification. We only considered the percentage of stained nuclei in each tumor, independent of the intensity. Three pathologists independently reviewed the results and those cases with discrepant results were discussed until agreement was achieved. As a negative control, we used non-immune rabbit serum instead of the KLF6 antibody.
The correlation of KLF6 expression with clinicopathologic parameters, such as tumor site, size, lymph node metastasis and clinical stage, was analyzed by χ2 test and Bartholomew test. P < 0.05 was considered statistically significant.
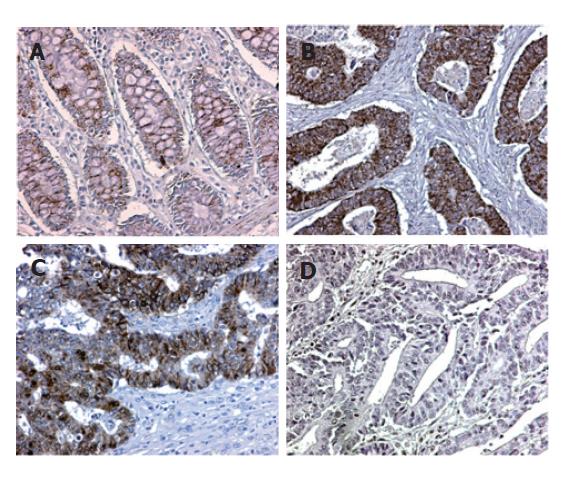
We assessed the pattern of KLF6 protein expression in 123 colorectal carcinoma specimens by immunohistochemistry using tissue microarray. The KLF6 expression was positive in cytoplasm and nuclei of normal colonic mucosal epithelial cells, but weak to moderate in lymphocytes and fibroblasts. The expression of KLF6 in tumor cells was compared to the expression in their corresponding normal colonic mucosae. Interestingly, reduced KLF6 expression or loss of KLF6 expression was found in 45 (36.6%) of the 123 colorectal carcinoma specimens, in which immunostaining on cytoplasm and nuclei of tumor cells was predominantly (Figure 1). Unexpectedly, over-expression of the KLF6 in cancer was also found in 12 cases. Negative immunoreactivity was seen in 15.4% (2 of 13 cases) of stage A, 38.3% (18 of 47) of stage B, 41.1% (23 of 56) of stage C, and 25.0% (2 of 8) of stage D, respectively (Table 1). Loss of expression was found in 25 (43.1%) of 58 cases with metastasis to regional lymph nodes and 32 (32.7%) of 98 left-side colorectal cancers. In addition, immunostaining was negative in 31 (47.0%) of 66 cases with tumors >5 cm. Statistically, loss of KLF6 expression was closely associated with tumor size (χ2 test, P < 0.05). However, there was no significant correlation between KLF6 expression and Dukes’ stage (Bartholomew test, P > 0.05), tumor location and lymph node metastasis (χ2 test, P > 0.05).
It has been reported that several KLFs play an essential role in maintaining the homeostasis of mammalian cells through the regulation of proliferation and differentiation[4,5]. The KLF6 is regulated during mammalian development and tissue regeneration, while its aberrant expression is associated with tumor formation. Recently, KLF6 has been identified as a tumor suppressor gene in several kinds of cancer because of reduced expression and its frequent genetic alterations, such as loss of heterozygosity and mutation, as well as functional suppression of cell proliferation. To further clarify the role of KLF6 in colorectal cancer, we examined loss of expression in 123 colorectal cancer samples.
In this study, reduced KLF6 expression or loss of KLF6 expression was found in 45 (36.6%) of the 123 colorectal adenocarcinoma specimens. Statistically, loss of KLF6 expression was closely associated with tumor size (χ2 test, P < 0.05). However, there was no significant correlation between reduced KLF6 expression and other clinicopathologic parameters, including Dukes’ stage (Bartholomew test, P > 0.05), tumor location and lymph node metastasis (χ2 test, P > 0.05). It has been reported that the expression of KLF6 protein is frequently down-regulated in non-small cell lung cancer and glioblastoma [7,14]. Interestingly, down-regulation of KLF6 expression is not associated with the histology or surgical-pathological stages of lung tumors[7]. Taken together, these results further support that down-regulation of KLF6 may contribute to the development of human solid cancers and that inactivation of KLF6 may occur in the development of colorectal cancer as an early event.
Unexpectedly, upregulation of KLF6 was also seen in 14 colorectal cancer cases. Since KLF6 is broadly involved in growth-related signal transduction, cell proliferation, differentiation and apoptosis, upregulation of KLF6 might reflect compensatory mechanisms directed at reestablishing the equilibrium in proliferation, differentiation, and survival characteristics of colonic epithelial cells disrupted during carcinogenesis. Because mutation of the KLF6 is frequently found in colorectal cancer [13], over-expression of KLF6 might be caused by the genetic alteration of the KLF6 gene. Further studies are necessary to investigate the precise molecular mechanisms underlying its over-expression.
Tissue microarray (TMA) consists of numerous tissues and tumor types. The use of TMA can stain a large number of cases at minimal cost and allow a rapid assessment of the staining profile. However, whether cores of tumor are representative of whole sections remains unclear. It was reported that immunostaining of one to four 0.6-mm cores is correlated with whole sections staining [19], suggesting that the use of small cores is equivalent to whole section immunohistochemistry. Another issue for consideration is that the scoring of TMA immunohistochemistry experiments remains subjective and not easily quantifiable. Significant efforts have been made to quantify protein expression levels on TMA sections [20], but most TMAs are subjectively scored by eyes. In this study, expression of KLF6 in three cores of tumor tissues was compared to their corresponding normal colonic mucosae by three pathologists. Although we did not perform immunohistochemistry on whole section, our results on TMA may be consistent with those of whole section immunohistochemistry.
Generally, the inactivation of a tumor suppressor associated with loss of expression results from genetic or epigenetic alterations, such as mutation, allelic loss or hypermethylation of the promoter region of the genes. Recently, genetic alterations of KLF6 gene have been identified in several human cancers, including hepatocellular carcinoma, prostate, gastric and colon cancers[10-13]. However, no mutation of KLF6 has been found in human cancer[21-23]. Further studies on a large number of patients are necessary to find out the discrepancy in incidence of KLF6 mutation. Interestingly, the KLF6 mRNA is also frequently down-regulated in non-small cell lung cancer, glioblastoma, and colon cancer cell lines[7,14]. No mutation has been found in these cancer tissues and KLF6 is down-regulated in all samples with allelic loss[7]. Additionally, the KLF6 is suggested to be a methylation-silenced gene in esophageal squamous cell carcinoma[24]. In the present study, we found that KLF6 expression was reduced or lost in 36.6% of colorectal cancers (Table 1). Although we did not perform the genetic or epigenetic analysis of the KLF6 gene, our results suggest that reduced KLF6 expression or loss of KLF6 expression may be a common and early event in colorectal carcinogenesis.
Co-first authors: Yong-Gu Cho and Byung-Jun Choi
S- Editor Wang J L- Editor Wang XL E- Editor Cao L
| 1. | Day DW, Jass JR, Price AB, Shepherd NA, Sloan JM, Talbot IC, Warren BF, Williams GT. Epithelial tumors of the large intestinge. In Morson BC, Dawson IMP, and Day DW. Morson and Dawson's gastrointestinal pathology. 4th ed. Oxford: Blackwell Pub 2003; 551-609 doi: 10.1002/9780470755983.ch38. |
| 2. | Shin HR, Jung KW, Won YJ, Park JG. 2002 annual report of the Korea Central Cancer Registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004;36:103-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 3. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8006] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 4. | Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355-34358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 494] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 5. | Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 850] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 6. | Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 352] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 7. | Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, Kawabe T, Hasegawa Y, Shimokata K, Sekido Y. Krüppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64:3838-3843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 126] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885-3891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 9. | Slavin DA, Koritschoner NP, Prieto CC, López-Díaz FJ, Chatton B, Bocco JL. A new role for the Kruppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene. 2004;23:8196-8205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson HF, Vessella RL, Dong JT. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol. 2003;162:1349-1354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 114] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P, Martignetti JA. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 112] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, Lee SH, Yoo NJ, Lee JY, Park WS. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588-4590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Kimmelman AC, Qiao RF, Narla G, Banno A, Lau N, Bos PD, Nuñez Rodriguez N, Liang BC, Guha A, Martignetti JA. Suppression of glioblastoma tumorigenicity by the Kruppel-like transcription factor KLF6. Oncogene. 2004;23:5077-5083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 15. | Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1474] [Cited by in RCA: 1341] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 16. | ASTLER VB, COLLER FA. The prognostic significance of direct extension of carcinoma of the colon and rectum. Ann Surg. 1954;139:846-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 757] [Cited by in RCA: 744] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 17. | Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2991] [Cited by in RCA: 2977] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 18. | Park WS, Oh RR, Kim YS, Park JY, Shin MS, Lee HK, Lee SH, Yoo NJ, Lee JY. Absence of mutations in the kinase domain of the Met gene and frequent expression of Met and HGF/SF protein in primary gastric carcinomas. APMIS. 2000;108:195-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Simon R, Mirlacher M, Sauter G. Tissue microarrays in cancer diagnosis. Expert Rev Mol Diagn. 2003;3:421-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 611] [Cited by in RCA: 619] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 21. | Lièvre A, Landi B, Côté JF, Veyrie N, Zucman-Rossi J, Berger A, Laurent-Puig P. Absence of mutation in the putative tumor-suppressor gene KLF6 in colorectal cancers. Oncogene. 2005;24:7253-7256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Koivisto PA, Hyytinen ER, Matikainen M, Tammela TL, Ikonen T, Schleutker J. Kruppel-like factor 6 germ-line mutations are infrequent in Finnish hereditary prostate cancer. J Urol. 2004;172:506-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Boyault S, Hérault A, Balabaud C, Zucman-Rossi J. Absence of KLF6 gene mutation in 71 hepatocellular carcinomas. Hepatology. 2005;41:681-62; author reply 681-62;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 261] [Article Influence: 11.3] [Reference Citation Analysis (0)] |