INTRODUCTION
Fibrosis is associated with many liver diseases, including hepatitis C virus infection, iron deposition, alcohol consumption, and nonalcoholic fatty liver disease. Hepatic fibrosis results from a net increased synthesis and decreased degradation of extracellular matrix (ECM) proteins. TypeIcollagen is the most prevalent ECM protein deposited[1], with activated hepatic stellate cells(HSCs) serving as the primary source. Following a fibrogenic stimulus, HSCs are activated from their normal quiescent state, whereby they increase synthesis of procollagen type I protein[1,2], and increase cellular proliferation, migration, and contractility[3,4]. Excess ECM accumulation results in scarring within the tissue[5].
In the progression of HSCs activation, many kinds of cytokines play important roles, including transforming growth factor-beta (TGF-β)[6]. TGF-β is a multifunctional cytokine with diverse effects on development, growth, and homeostasis in most tissues[7]. Signaling by TGF-β occurs through typeIand typeIItransmembrane serine/threonine kinase receptors and intracellular Smad transduction molecules. The TGF-β ligand binds to its type II receptor, either directly or via coreceptors. Once activated by TGF-β, type II receptors recruit, bind, and transphosphorylate type I receptors, thereby stimulating protein kinase activity of the later. The activated type I receptors phosphorylate Smad2 or Smad3, which then bind to Smad4. The resulting Smad complex then moves into the nucleus, where it interacts in a cell-specific manner with various transcription factors to regulate the transcription of target genes[8].
It has been reported that there is a correlation between TGF-β and the formation of liver fibrosis[9,10]. TGF-β has a very important role in ECM production by HSC and many investigations have been done[11,12]. Antagonism of cytokines as a route to antifibrotic therapy, by blocking the interaction between TGF-β and its receptors, and the signal transduction mechanism for TGF-β, has been reported for the treatment of fibrosis[13,14]. Thus the investigation of the Smads signal transduction is indispensable for the study of fibrosis and the development of its therapy. Until now, there have been no reports on the location and expression alteration of Smad2,4 during and after fibrogenesis. In this article, the expression and location of Smad2,4 mRNA were measured in rat livers with cRNA probes of Smad2,4 by in situ hybridization.
MATERIALS AND METHODS
Animal experiments
Eighty male Wistar rats weighing approximately 200 g each were used. Chronic liver injury was produced in 10 rats in each group by subcutaneous injections of 50% CCl4 in olive oil at a volume of 2 ml/kg body weight twice weekly for 3, 6, 9, 12, 15, and 18 wk. Ten control rats similarly received olive oil alone as control group, and 10 other rats were not treated as normal group. All rats were maintained on a standard diet with water ad libitum. On the sixth day after the last treatment, 10 rats in each group were decapitated. Their livers were removed immediately, fixed in 4% paraformaldehyde at room temperature and snap-frozen in liquid nitrogen. The tissue specimens were stored at -80 °C until analysis.
Cloning of Smad2 and Smad4 cDNAs
Smad2 and Smad4 cDNAs were cloned in order to synthesize ISH riboprobes. The two cDNAs were obtained from rat liver total RNA by reverse transcription coupled to PCR by the use of the primers Smad2-up (5’GGTGGATCCGTGTCTCATCGGAAAGGG-3’), Smad2-GTGAATTCTGGAATGGAGTGGGTATAG-3’) and Smad4-up (5’-AACCGGATCCATCTTCAGC ACCACCC-3’), Smad4-down (5’CGAATTCTTTGCCTATGTGCAACCTTGC-3’), respectively. The two resulting fragments were digested with BamHIand EcoRIand cloned into the pSPT18 vector (Boehringer Mannheim) to obtain the pSPT18+/Smad2 and pSPT18+/Smad4 plasmids.
ISH riboprobe synthesis
The cDNA-containing plasmids were linearized as follows: pSPT18+/Smad2 and pSPT18+/Smad4 with BamH I and EcoRI. The linearized plasmids were then gel-isolated (Hua Shun Corp., China) and used as templates for anti-sense and sense digoxigenin (DIG)-labeled riboprobe synthesis(Boehringer Mannheim). The transcription mixture (20 µL) included 1 µg of linearized template cDNA , ATP, GTP and CTP at 1 mmol/L each, UTP 0.7 mmol/L, DIG-UTP 0.3 mmol/L, DTT 10 mmol/L, RNase inhibitor (1 U/µL of transcription mix), and T3 or T7 RNA polymerase (1 U/µL of transcription mix). Transcription was performed for at least 2 hr at 37 °C. The template cDNAs were then digested by RNase-free DNase (2 µL at 1 U/µL, 15 min at 37 °C), and all reactions were stopped by adjusting the reaction volume to 100 µL with EDTA (pH8.0). The riboprobes were then purified through two precipitation steps by addition of 100 µL LiCl (4 mmol/L) and 500 µL EtOH (100%), and centrifugation for 30 min at 4 °C in a microfuge. The pellet was resuspended in 100 µL DECP-treated water.
In situ hybridization
The serial paraffin sections (thickness 4 µm) were dried at 55 °C, then deparaffinized by xylene and rehydrated in graded ethanol, incubated for 2×5 min in TBS containing 0.1% active DEPC (Fluka), acidified in 0.2 N HCl for 20 min at room temperature, and incubated for 2×5 min in TBS before digestion with proteinase K for 15 min, and then incubated for 2×5 min in TBS. After postfixation in 4% paraformaldehyde-PBS, sections were incubated for 2×5 min in TBS. And then sections were dehydrated with graded ethanol. After prehybridization at 42 °C for 2 h, the labeled RNA probes were added to the hybridization mix (Smad2 3.7 ng/µL, Smad4 2.3 ng/µL). The hybridization was carried out at 42 °C for 18 h with 20µL of hybridization mix on each section. The sections were washed with 2×SSC, 1×SSC, and TBST(TBS containing Tween20) respectively. After washed, the sections were incubated for 15 min at room temperature with Buffer I containing 1% blocking reagent (Boehringer Mannheim), and then at 37 °C with alkaline phosphatase-coupled anti-digoxigenin antibody(Boehringer Mannheim) diluted at 1:50 in Buffer I. Excess antibody was removed by 5×4 min washes in TBST, and the sections were equilibrated for 2×5 min in Buffer I(Tris-HCl 100mmol/L, NaCl 100mmol/L, and MgCl2 100mmol/L, pH9.5). Color development was performed at 37 °C for 8 h in Buffer I containing NBT and BCIP(Promega). Staining was stopped by a 10 min washing in tap water, and non-special staining was removed in EtOH 95% with gentle agitation. Then the sections were rehydrated through successive baths of EtOH (70, 95, and 100%) and xylol (2×15 min each) and mounted in gum for microscopic examination and photography. Blank control: Smad2 and Smad4 cRNA probes for positive hepatic tissues were replaced by prehybridization solution. Negative control: in situ hybridization was performed in normal liver tissues. False positive control: in situ hybridization was performed in liver tissues injected with olive oil.
Pathology observation
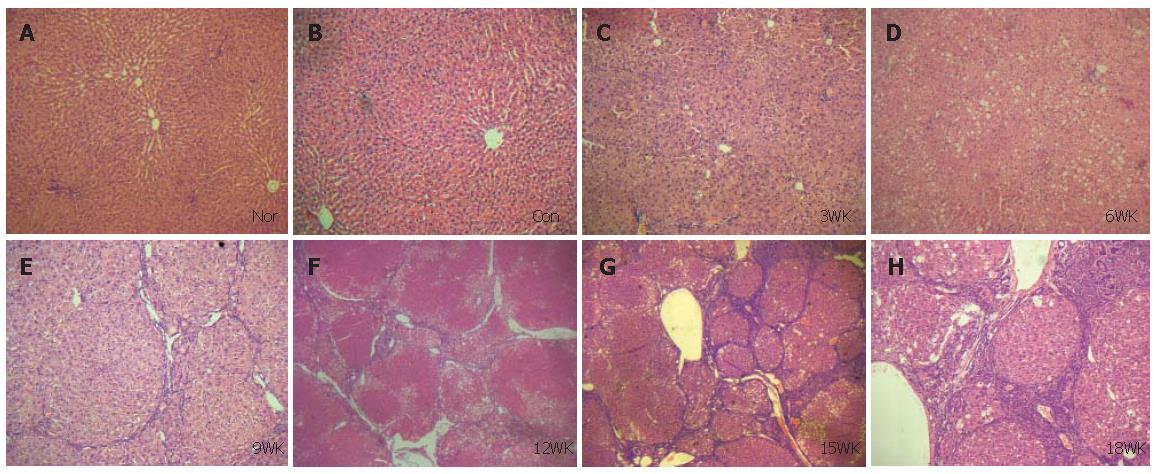
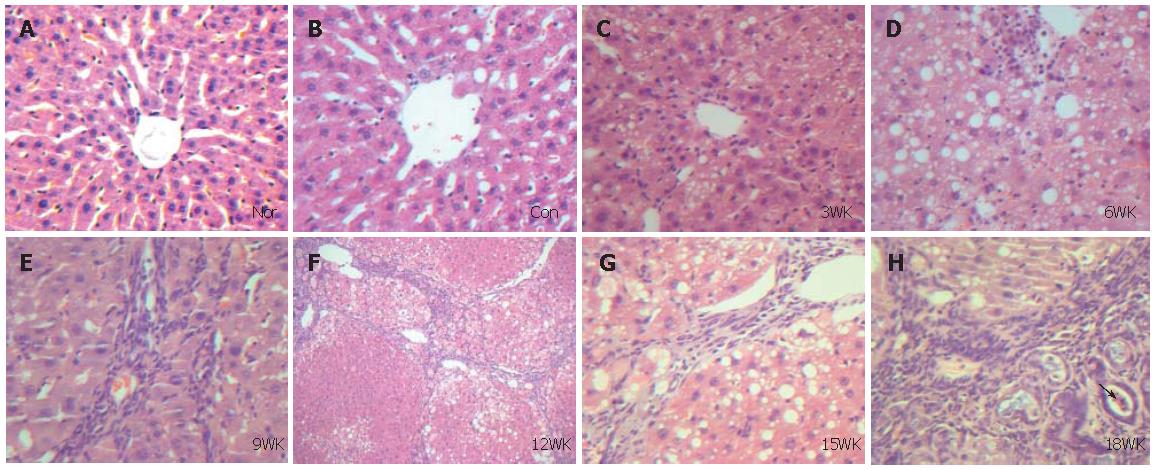
The whole liver sections were stained with hematoxylin and eosin, and examined under microscope.
Statistic analysis
The t test was used to determine statistical significance between groups. P value of less than 0.05 was considered statistically significant.
DISCUSSION
To establish the model of liver fibrosis successfully is the key for detection of Smad2 and Smad4 mRNAs in the liver tissue during and after fibrogenesis. Researches for establishing the model of liver fibrosis with CCl4 began in 1936. After that many methods to establish the model of liver fibrosis have been tried. Among them, liver fibrosis models induced by immune response or CCl4 have been generally accepted mainly because they are more close to the disease characteristics in terms of the distinct stages of the disease of human body and a low mortality. In this study, the method of injecting CCl4 was used to establish the model of liver fibrosis, and it was proved successful by histochemical analysis of the sections stained with hematoxylin and eosin. According to the pathologic findings of this study, the exact time for establishment of rat hepatic cirrhosis by injecting CCl4 may be at 12 wk. From 15 and 18 wk on, atypical hyperplasia was found in the epithelium of bile capillary. It may indicate that injecting CCl4 may cause hepatic carcinoma.
TGF-β consists of a big molecular family that has a common structure including BMP and activin. These proteins have many roles in the cell structure and function, such as cell growth, differentiation, apoptosis and regulation of ECM production[15,16]. The main functions of the TGF-β super family are morphogenesis, inflammation, tissue recovery and oncogenesis[17]. Smads are intracellular signal transductive molecules of the TGF-β super family. These molecules were discovered as Mothers against dpp (Mad)[18,19], or sma[20] and named Smad in 1996[21]. According to the differences of structure and function, nine Smads have been reported and classified into three groups[7,22]. Smads 2 and 3 are named R-Smads in the pathway and Smad 4 a Co-Smads for all these pathways. Smads 6, 7, 8 and Xsmad 8 are inhibitory factors of these Smads. When TGF-β binds to its receptor, Smad 2/3 is phosphorylated and binds with Smad 4, together they move into the nucleus for translation and expression of the target gene[7,23]. These mechanisms are thought to play a role in the process of liver damage, recovery, as well as liver fibrosis. In a damaged liver including chronic inflammation, in hepatitis B, TGF-β signaling was reported[24]. Not only Kupffer cells but also the HSC produce TGF-β by the autocrine system[25-28]. TGF-β1 stimulation evokes ECM production, which develops into liver fibrosis.
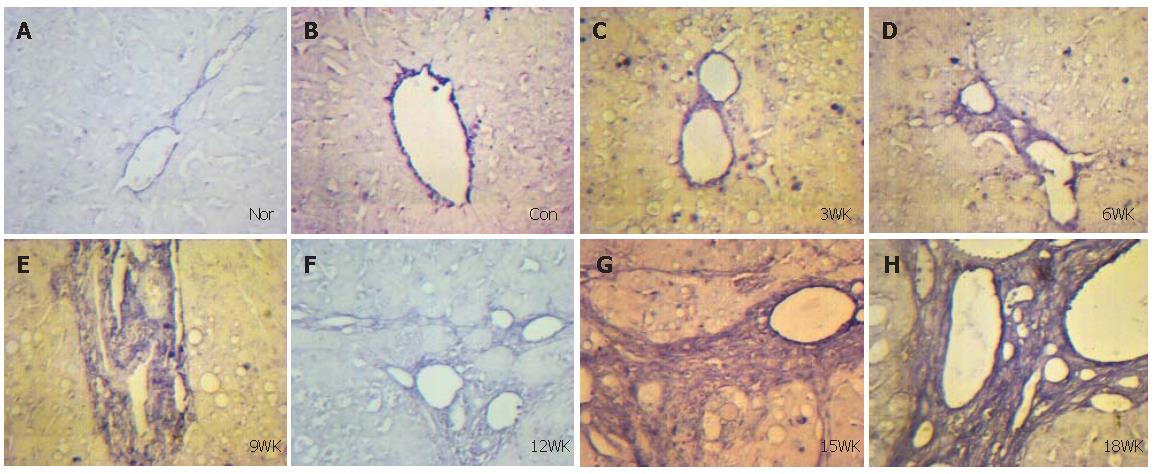
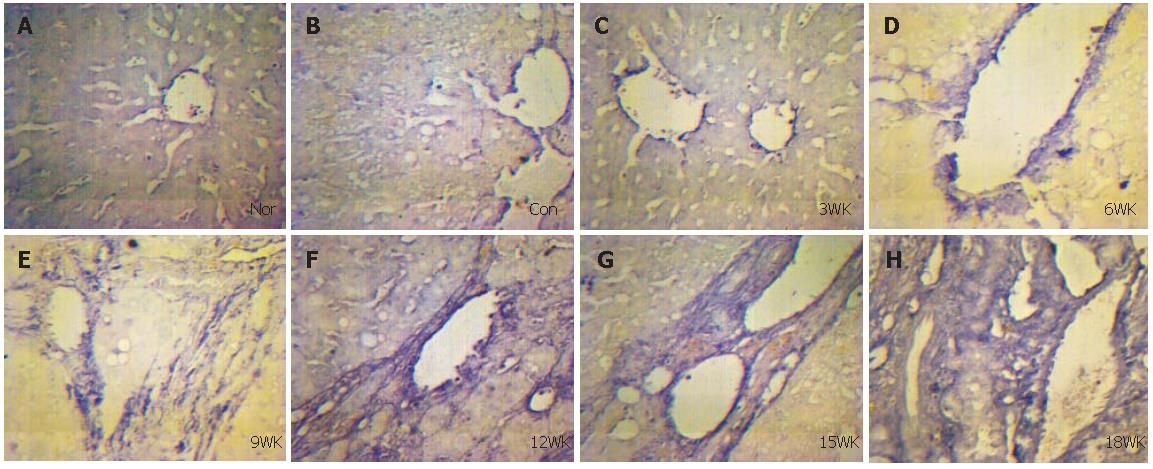
In this study, Smad2 and Smad4 mRNAs were investigated in rat liver during and after fibrogenesis caused by CCl4in vivo. With the method of in situ hybridization, we found Smad2 and Smad4 mRNAs were expressed obviously in HSC, myofibroblasts and fibroblasts of livers of liver fibrosis group, mostly in portal area and fibrous septum, while very mildly in vascular endothelial cells of livers from normal and control groups. While with the development of liver fibrosis, expressions of Smad2 and Smad4 mRNAs were increased also. These results indicate that Smad2 and Smad4 play an important role in the progression of liver fibrosis. In the sections of liver fibrosis at 15 and 18 wk, expressions of Smad2 and Smad4 mRNAs were higher in the epithelium of bile capillary than in normal and control groups. At this stage, there was atypical hyperplasia in the epithelium of bile capillary, indicating early stage of carcinoma. Therefore, Smad2 and Smad4 may be associated with cancerogenesis.
After stimulation of TGF-β, Smad2, 3 and 4 proteins translocate from cytoplasm to nucleus in MV1Lu cells, analyzed by immunofluorescence using specific antiserum[22]. Moreover, Smad3 seems to play the most important role in liver fibrosis, because Smad3 interacts with SP1 which increases expression of ECM by regulating COL1A2 gene[29]. By transfection of Smad3 expression plasmid in CF37 cells, ECM also increases by regulating COL1A2 gene. Nevertheless, COL1A2 transcription in CF37 cells is markedly increased by TGF-β after transfection with a Smad2 expression plasmid[30]. So Smad2 also plays an important role in liver fibrosis. Our findings are consistent with the above results. The difference is perhaps due to use of different cell lines or experimental conditions. Further researches are needed to confirm the results.
As the liver fibrosis is a complex process, involving many cytokines, cells, and signal transduction pathways, the mechanism cannot be fully explained by these experiments. However, it is clear that TGF/Smads signal transduction pathway plays an important role in liver fibrosis which has been proved by many researches. In our study, Smad2 and Smad4 mRNAs were detected in the plasma of HSC, fibroblasts and myofibroblasts around the portal area and central vein by in situ hybridization. Expressions of Smad2 and Smad4 mRNAs were higher during and after liver fibrogenesis than normal and control groups, indicating that they play an important role in liver fibrosis.