Published online Mar 7, 2005. doi: 10.3748/wjg.v11.i9.1273
Revised: March 18, 2004
Accepted: March 24, 2004
Published online: March 7, 2005
AIM: To develop a microarray-based prewarning system consisting of gastric cancer chip, prewarning data and analysis software for early detection of gastric cancer and pre-cancerous lesions.
METHODS: Two high-density chips with 8 464 human cDNA sites were used to primarily identify potential genes specific for normal gastric mucosa, pre-cancerous lesion and gastric cancer. The low-density chips, composed of selected genes associated with normal gastric mucosa, precancerous lesion and gastric cancer, were fabricated and used to screen 150 specimens including 60 specimens of gastric cancer, 60 of pre-cancerous tissues and 30 of normal gastric mucosa. CAD software was used to screen out the relevant genes and their critical threshold values of expression levels distinguishing normal mucosa from pre-cancerous lesion and cancer. All data were stored in a computer database to establish a prewarning data library for gastric cancer. Two potential markers brcaa1 and ndr1 were identified by Western blot and immunohistochemistry.
RESULTS: A total of 412 genes associated with three stages of gastric cancer development were identified. There were 216 genes displaying higher expression in gastric cancer, 85 genes displaying higher expression in pre-cancerous lesion and 88 genes displaying higher expression in normal gastric mucosa. Also 15 genes associated with metastasis of gastric cancer and 8 genes associated with risk factors were screened out for target genes of diagnosis chip of early gastric cancer. The threshold values of 412 selected genes to distinguish gastric cancer, pre-cancerous lesion from normal gastric mucosa were defined as 6.01±2.40, 4.86±1.94 and 5.42±2.17, respectively. These selected 412 genes and critical threshold values were compiled into an analysis software, which can automatically provide reports by analyzing the results of 412 genes obtained by examining gastric tissues. All data were compiled into a prewarning database for gastric cancer by CGO software. Northern blot and immunohistochemistry analysis confirmed that gene and protein of brcaa1 displayed lower expression in normal gastric mucosa and higher expression in gastric cancer tissues, conversely, ndr1 displayed lower expression in gastric cancer and higher expression in normal gastric mucosa.
CONCLUSION: The microarray-based prewarning system for gastric cancer was developed. This system consisted of gastric cancer-associated gene chip, prewarning data and analysis software, which has a high potential for applications in the early detection of gastric cancer. The two potential markers brcaa1 and ndr1 identified may be used to distinguish cancer status fand non-cancer status.
- Citation: Cui DX, Zhang L, Yan XJ, Zhang LX, Xu JR, Guo YH, Jin GQ, Gomez G, Li D, Zhao JR, Han FC, Zhang J, Hu JL, Fan DM, Gao HJ. A microarray-based gastric carcinoma prewarning system. World J Gastroenterol 2005; 11(9): 1273-1282
- URL: https://www.wjgnet.com/1007-9327/full/v11/i9/1273.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i9.1273
Gastric cancer has high incidence in China and in the whole world. Understanding the biological processes of cancer initiation at the gene expression level is very important for early cancer detection. Study of gene expression levels at different stages of growth, disease, cell cycle, and response to stimulation may help to answer why different stages of cancerous development occur[1]. We have been trying to establish a prewarning system of gastric cancer as a part of a larger effort to develop effective and economical diagnostic tools capable of distinguishing different stages of cancer development. This system consists of three important parts: a gastric cancer microarray, a prewarning data library and a data analysis software.
Screening characteristic differentially expressed genes associated with different stages of cancer development is of central significance to this study. In our previous studies[2,3], some differentially expressed genes between gastric cancer tissues and precancerous lesions have been obtained. Genes that have been shown to correlate with gastric cancer were used as a part of the target genes in the microarray. Commercially available microarrays with 8 464 human cDNA sites have also been used for identifying specific genes associated with normal mucosa, precancerous lesions and gastric cancer.
The gene microarray technique has the advantage of simultaneously monitoring the expression of thousands of genes in one hybridization experiment. This technique has greatly facilitated the detection of differentially expressed genes and the construction of gene expression profiles. Since 1995, the DNA microarray technique has been widely employed to investigate the functions of genes, especially those genes involved in tumor generation and growth[4]. This technique has a great potential as a practical clinical tool for medical diagnosis[5]. Although many genes are known to be related to the pathological process of gastric carcinoma, so far very few prognostic biomarkers of gastric cancer have actually been used in clinical medicine. In our present study, we tried to identify specific genes involved in gastric carcinogenesis, with the objective of establishing a prewarning system for early diagnosis, therapy and prevention of gastric cancer.
Specimens used in this study were classified into three different categories: those of gastric cancer (including all types of pathologic gastric cancers such as diffuse type and intestinal type), those of paracancerous lesions (according to international classified standard including atrophic gastritis, intestinal gland metaplasia, atypical hyperplasia) and those of normal gastric mucosa (including slight superficial gastritis). A total of 150 specimens including 60 gastric cancers, 60 pre-cancerous lesions and 30 normal gastric mucosa in liquid nitrogen with clear pathological results, were provided by the department of Gastroenterology of the Xi’an Central Hospital. Human cDNA microarrays with 8464 were purchased from BioDao Company in Shanghai.
Total RNA was extracted by using total RNA extract kit from Promega Inc. Reverse transcriptionof mRNA was performed by using Smart PCR cDNA synthesis kit (Clontech). Reaction products were purified with Wizard plus minipreps DNA purification system. Cy3-dUTP, Cy5-dUTP and CSS-25 silylated slides (aldehyde) were purchased from Pharmacia Inc. and Gene Limited Inc. Spot reportTM oligoTMarray validation system (Cat # 252170-7) was purchased from Stratageneâ Company. Other reagents were purchased from Sigma Inc.
Microarrays consisting of 2435 fragment sites including 412 genes were fabricated. These synthesized oligonucleotide DNAs were first dissolved in 3×SSC solution. Spot report oligo array validation system (Cat # 252170-7) was used as quality control. Spots with pure 3×SSC solution were selected as background control. The target genes were spotted on silylated slides by MicroGridII spotting robotics (BioRotics Inc.). After spotting, the slides were hydrated (2 h), dried (0.5 h, RT), UV crosslinked (65 mJ/cm), and then treated with 2 g/L SDS (10 min), H2O (10 min), and 2 g/L NaBH4 (10 min). The slides were dried before being made ready for usage.
Total RNA extraction was performed by using total RNA extract kit from Promega Inc. Final total RNA templates were dissolved with non-RNase and non-DNase Milli-Q H2O. Fluorescent cRNA probes were prepared through reverse transcription and then purified, referring to the protocol of Schena(DNA microarrays, a practical approach. Oxford University Press, 1999:110-126). The probes from gastric cancer tissues and pre-cancerous tissues were labeled with Cy5-dUTP, those from normal gastric mucosa tissues with Cy3-dUTP. The labeled probes were mixed, fragmented and precipitated by ethanol and dissolved in 20 μL hybridization solution (5×SSC+2 g/L SDS).
After denatured at 95 °C for 5 min, the probes were added onto slides, covered with a cover glass and incubated at 42 °C for 17 h. The slides were subsequently washed in solutions of 2×SSC+2 g/L SDS, 0.1×SSC+2 g/L SDS and 0.1×SSC, 10 min each time, and then dried at room temperature.
Microarrays were scanned by using Affymetrixâ 428TM array scanner. ImageGene 3.0 software (BioDiscovery Inc.) was used to quantify, correct for background noise and normalize the signals from post-hybridization chip.
The data files were incorporated into a computer database by CGO software, including patient disease history and all screened results, such as, name, file number, sex, age, address, telephone, e-mail address, marital status, blood type, body mass, disease history, imaging examination, pathological examination, serum examination, blood examination, cytogenetic report, and gene array report.
Expression gene profiles were established according to the acquired data. CAD software was used in the selection of discriminating candidate genes by their correlation with three kinds of gastric tissues, determination of the optimal set of reporter genes by using a leave-one-out validation procedure, determination of the threshold values of selected gene expression levels to distinguish normal gastric mucosa from pre-cancerous lesions and gastric cancer, and metastatic cancer and no-metastatic cancer.
A total of 412 genes and critical threshold values to distinguish normal gastric mucosa from pre-cancerous lesion and gastric cancer were compiled into an analysis software, which could provide analysis reports by analyzing the microarray test results.
Five micrograms of mRNA was resolved by denaturing formaldehyde agarose gel and transferred onto hybrid membranes (Amersham). The membranes were hybridized with 32p-labeled fragments of cDNA overnight, washed twice in 1 g/L standard saline citrate and 1 g/L SDS for 20 min and then exposed to Kodak BioMax film at -80 °C with an intensifying screen for 24 h.
Standard avidin-biotin complex (ABC) technique was used for immunohistochemical staining of formalin-fixed, paraffin-embedded gastric cancer tissues. Specific antibody (10 mg/L) and PBS were added onto tissue slides previously blocked with rabbit serum and incubated overnight. After washing with PBS, the slides were incubated with a rabbit anti-human IgG conjugated to biotin at room temperature for 1 h, alkaline phosphatase substrate was then added for color development. The slides were counterstained with hematoxylin-eosin.
A two-way clustering analysis was performed by using Cluster software and Tree view software from http://www.microarray.org(PNAS 1998; 95:14863). Statistical analysis was performed by using the t test. All P values were based on two-sided testing, and a significant difference was defined as P less than 0.05.
Two high-density chips were used to primarily screen differential genes associated with normal gastric mucosa, pre-cancerous lesion and gastric cancer. According to the obtained partial biochip hybridization results, 393 genes closely associated with three stages of gastric cancer development were primarily screened out (Figure 1). Fifteen genes associated with gastric cancer metastasis and 8 genes associated with risk factor genes of gastric cancer, such as cagA, vacA, Ure, EB, were selected according to the literature[6]. These genes were used as main target genes on the prewarning chip. The oligonucleotides associated with 412 genes were designed, synthesized and fabricated into low-density chip.
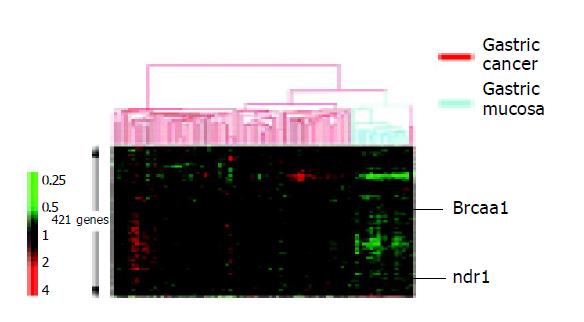
All the 150 specimens with clear pathological results were screened with the fabricated low-density microarrays. Among these, 60 were known to be cancerous, 60 precancerous and 30 normal (Figure 2). In the 60 cancer specimens, 216 genes were found to exhibit higher expression levels than those in normal gastric mucosa. Among the 216 genes, 156 also exhibited higher expression levels than those in the precancerous lesions (Table 1). In the 60 specimens of Pre-cancerous lesions, 126 genes exhibited higher expression levels than those in the normal tissues. Among those, 85 genes also showed higher expression levels than those in the gastric cancer tissues (Table 1). Contrary to our initial expectations, selected risk factor genes such as cagA, vacA, Ure, EB did not show overexpression levels in gastric cancer tissues in comparison with the normal tissues and precancerous lesions. In fact, these genes showed lower expression levels in gastric cancer tissues than in normal tissues and precancerous lesions. This result demonstrated that the risk factor genes due to H pylori infection might be more closely associated with the progression of precancerous lesion. Eighty-eight genes in normal tissues exhibited higher expression levels than those found in gastric cancer tissues and pre-cancerous tissues (Table 1). These genes are helpful for distinguishing normal gastric mucosa from precancerous lesions. This is very important in diagnosing the precancerous lesion among common gastric diseases, such as superficial gastritis, because the treatment of precancerous lesion requires special focused methods. If left untreated, precancerous lesion might result in gastric cancer in a limited time.
| GenBank | Number | Description of gene |
| Highly expressed genes in gastric cancer | ||
| 1 | NM_001962 | Homo sapiens ephrin-A5 (EFNA5) |
| 2 | XM_017384 | Homo sapiens matrix metalloproteinase 7 (MMP7) |
| 3 | NM_008610 | Mus musculus matrix metalloproteinase 2 (Mmp2) |
| 4 | NM_004995 | Homo sapiens matrix metalloproteinase 14 (MMP14) |
| 5 | AF093573 | Bos taurus angiopoietin-1 (ang-1) |
| 6 | AF004327 | Homo sapiens angiopoietin-2 |
| 7 | M11730 | Human tyrosine kinase-type receptor (HER2) |
| 8 | U13948 | Human zinc finger/leucine zipper protein (AF10) |
| 9 | XM_049646 | Homo sapiens similar to octamer-binding transcription factor 3B (OCT-3B) |
| 10 | XM_055784 | Homo sapiens fibroblast growth factor 2 (basic) (FGF2) |
| 11 | XM_056035 | Homo sapiens proliferating cell nuclear antigen (PCNA) |
| 12 | L24203 | Homo sapiens ataxia-telangiectasia group D-associated protein |
| 13 | XM_087201 | Homo sapiens similar to RED protein, IK cytokine |
| 14 | X00663 | Human mRNA fragment for epidermal growth factor (EGF) receptor |
| 15 | NM_002607 | Homo sapiens platelet-derived growth factor alpha polypeptide (PDGFA) |
| 16 | XM_165656 | Homo sapiens matrix metalloproteinase 2 (MMP2) |
| 17 | NM_005918 | Homo sapiens malate dehydrogenase 2, NAD (mitochondrial) (MDH2) |
| 18 | AF503165 | Homo sapiens HUS1 checkpoint homolog (HUS1) gene |
| 19 | XM_045667 | Homo sapiens antigen identified by monoclonal antibody Ki-67 (MKI67) |
| 20 | XM_050913 | Homo sapiens frequently rearranged in advanced T-cell lymphomas (FRAT1) |
| 21 | XM_032866 | Homo sapiens signal transducer and activator of transcription 5A (STAT5A) |
| 22 | NM_004103 | Homo sapiens protein tyrosine kinase 2 beta (PTK2B) |
| 23 | XM_008355 | Homo sapiens membrane protein, palmitoylated 2 (MPP2) |
| 24 | L18920 | Human MAGE-2 gene exon 2, 3, 4 |
| 25 | M12174 | Human ras-related rho |
| 26 | NM_012333 | Homo sapiens c-myc binding protein (MYCBP) |
| 27 | BC016514 | Homo sapiens, similar to translocated promoter region (to activated MET oncogene) |
| 28 | NM_004324 | Homo sapiens BCL-2 associated X protein (BAX) |
| 29 | Z26580 | cyclin A |
| 30 | D45906 | LIMK-2 |
| 31 | D21255 | OB-cadherin-2 |
| 32 | X54925 | Type I interstitial collagenase |
| 33 | X05232 | Stromelysin, matrix metalloproteinase 3 |
| 34 | M22612 | Human pancreatic trypsin 1 (TRY1) |
| 35 | XM_055254 | Homo sapiens fibronectin 1 (FN1) |
| 36 | AF081127 | Danio rerio fibronectin (fn2) |
| 37 | M15796 | Human cyclin protein gene |
| 38 | HSFIBEDA | Human fibronectin gene ED-A region |
| 39 | HSU66406 | Human putative EPH-related PTK receptor ligand LERK-8 (Eplg8) |
| 40 | AF068846 | Homo sapiens scaffold attachment factor A (SAF-A) |
| 41 | HSBTRCP | Homo sapiens mRNA for beta-transducin repeat containing protein |
| 42 | AF110763 | Homo sapiens skeletal muscle LIM-protein 1 (FHL1) gene |
| 43 | HUMHO2SOS1 | Human mRNA for heme oxygenase-2 |
| 44 | HSHMSH16 | Human mutator hMSH2 gene |
| 45 | HSEHK1 | Homo sapiens mRNA for EHK-1 receptor tyrosine kinase |
| 46 | HSKLON30 | Homo sapiens mRNA for unknown antigen |
| 47 | AB005047 | Homo sapiens mRNA for SH3 binding protein |
| 48 | AF070561 | Homo sapiens clone 24703 beta-tubulin |
| 49 | HUMCAM1V | Human vascular cell adhesion molecule 1 |
| 50 | HSRNASMG | Homo sapiens mRNA for Sm protein G |
| 51 | X83228 | Homo sapiens mRNA for LI-cadherin |
| 52 | AF125100 | Homo sapiens HSPC039 protein |
| 53 | HSU97018 | Homo sapiens echinoderm microtubule-associated protein homolog HuEMAP |
| 54 | HSU43188 | Human Ets transcription factor (NERF-2) |
| 55 | HSY17392 | Homo sapiens mRNA for prefoldin subunit 1 |
| 56 | HSU08316 | Human insulin-stimulated protein kinase 1 (ISPK-1) |
| 57 | HZNF232G2 | Homo sapiens zinc finger protein ZNF232, exons2 and 3 |
| 58 | HUMP53T | Human p53 cellular tumor antigen |
| 59 | J03040 | Human SPARC/osteonectin |
| 60 | XM_053809 | Homo sapiens similar to chondroitin sulfate proteoglycan 2 (versican) |
| 61 | L40379 | Homo sapiens thyroid receptor interactor (TRIP10) |
| 62 | HSU72069 | Human karyopherin beta2 |
| 63 | HUMPGK2 | Human phosphoglycerate kinase (pgk) mRNA, exons 2 to last |
| 64 | HSU07139 | Human voltage-gated calcium channel beta subunit |
| 65 | XM_001472 | Homo sapiens v-jun sarcoma virus 17 oncogene homolog (avian) (JUN) |
| 66 | AU100088 | Human phosphogluconate dehydrogenase (hPGDH) gene |
| 67 | HUMKRUPZN | Human Kruppel related zinc finger protein (HTF10) |
| 68 | AF077050 | Homo sapiens neuroendocrine-specific protein C homolog |
| 69 | HUMSC35A | Human splicing factor SC35 |
| 70 | HUMPTPB | Homo sapiens protein tyrosine phosphatase (CIP2) |
| 71 | AF049608 | Homo sapiens monocarboxylate transporter 2 (MCT2) |
| 72 | HUMHEK | Human receptor tyrosine kinase (HEK) |
| 73 | J03210 | Human collagenase type IV |
| 74 | HSRAB9P40 | Homo sapiens mRNA for Rab9 effector p40 |
| 75 | AF184924 | Homo sapiens zinc finger transcription factorBTEB2 gene |
| 76 | HUMC5A2A | Human fibrillar collagen (proa2 (V)) gene |
| 77 | HUMGAPA | Human GTPase-activating protein ras p21 (RASA) |
| 78 | HUMGLURS | Human glutamate receptor subunit (GluH1) |
| 79 | AF047715 | Homo sapiens A-kinase anchoring protein (AKAP18) |
| 80 | HSU40282 | Homo sapiens integrin-linked kinase (ILK) |
| 81 | HSATPF1M | Human mRNA for mitochondrial ATP synthase(F1-ATPase) alpha subunit |
| 82 | AF152485 | Homo sapiens protocadherin alpha 7 short formprotein (PCDH-alpha7) |
| 83 | HSRP19 | Human mRNA for 19 ku protein of signal recognition particle (SRP) |
| 84 | U17195 | Homo sapiens A-kinase anchor protein (AKAP100) |
| 85 | HSU79299 | Human neuronal olfactomedin-related ER localizedprotein |
| 86 | XM_037859 | Human focal adhesion kinase (FAK) |
| 87 | HSU04209 | Human-associated microfibrillar protein |
| 88 | D82878 | Hemicentrotus pulcherrimus mRNA for p34cdc2 |
| 89 | AF060515 | Homo sapiens cyclin K (CPR4) |
| 90 | D21262 | Human mRNA for KIAA0035 gene |
| 91 | NM_005641 | Homo sapiens TATA box binding protein-associatedfactor, RNA polymerase II, 85 ku |
| 92 | HSU07550 | Human chaperonin 10 |
| 93 | X82153 | Homo sapiens mRNA for cathepsin 0 |
| 94 | HSU41766 | Human metalloprotease/disintegrin/cysteine-richprotein precursor (MDC9) |
| 95 | AB017019 | Homo sapiens mRNA for JKTBP2 |
| 96 | HUMFNC | Human cellular fibronectin |
| 97 | U93033 | Homo sapiens thyroglobulin (TG) |
| 98 | AF0304354 | Homo sapiens proteoglycan 3 (PRG3) gene |
| 99 | HUMCOL3IX | Homo sapiens collagen alpha 3 type IX (COL9A3) |
| 100 | NM_002427 | Homo sapiens matrix metalloproteinase 13(MMP13) |
| 101 | AF039747 | Homo sapiens cadherin-10 (CDH10) |
| 102 | AF072242 | Homo sapiens methyl-CpG binding protein MBD2(MBD2) |
| 103 | HSMYCC | Human c-myc oncogene |
| 104 | HSTSPM | Homo sapiens tissue specific mRNA |
| 105 | HSU64317 | Human Crk-associated substrate relatedprotein Cas-L |
| 106 | HSVACM1 | Homo sapiens mRNA for vasopressin activatedcalcium mobilizing receptor-like protein |
| 107 | HUMPA1V | Human pro-alpha-1 (V) collagen |
| 108 | AF059611 | Homo sapiens nuclear matrix protein NRP/B (NRPB) |
| 109 | HSU004845 | Human a6 (I V) collagen (COL4A6) |
| 110 | M87860 | Human S-lac lectin L-14-II (LGALS2) gene |
| 111 | AF492837 | Human mRNA for osteopontin |
| 112 | HSCOX7BM | Homo sapiens coxVIIb mRNA for cytochromec oxidase subunit VIIb |
| 113 | U01244 | Human fibulin-1D |
| 114 | U52153 | Human inwardly rectifying potassium channelKir3.2 |
| 115 | S66427 | RBP1=retinoblastoma binding protein 1 [human, Nalm-6 pre-B cell leukemia, mRNA, 4834 nt] |
| 116 | AF117108 | Homo sapiens IGF-II mRNA-binding protein3 (IMP-3) |
| 117 | HSU49083 | Human cell surface heparin binding protein HIP |
| 118 | HSU59289 | Human H-cadherin |
| 119 | HSU95032 | Human growth-arrest-specific protein 2 |
| 120 | HSU18018 | Human E1A enhancer binding protein (E1A-F) |
| 121 | HUMCGRPB | Homo sapiens (clone HSNME29) CGRP type1 receptor |
| 122 | X59543 | Human mRNA for M1 subunit of ribonucleotidereductase |
| 123 | AF072810 | Homo sapiens transcription factor WSTF |
| 124 | AF005068 | Homo sapiens breast and ovarian cancersusceptibility protein splice variant (BRCA1) |
| 125 | HSU66197 | Human fibroblast growth factor homologous factor1 (FHF-1) |
| 126 | HUMVTNR | Human cell adhesion protein (vitronectin) receptoralpha subunit |
| 127 | HSA6417 | Homo sapiens mRNA for beta-tubulin foldingcofactor D |
| 128 | AF109126 | Homo sapiens stromal cell-derived receptor-1 beta |
| 129 | AB030078 | Homo sapiens mRNA for K-sam-II03 |
| 130 | HUMMFAP | Homo sapiens extracellular matrix protein (MFAP3)gene |
| 131 | HUMCOLVA | Human alpha-2 type V collagen gene |
| 132 | HUMAAMP1X | Homo sapiens angio-associated migratory cell protein (AAMP) |
| 133 | Y08319 | Homo sapiens mRNA for kinesin-2 |
| 134 | HSVWFR1 | Human mRNA for pre-pro-von Willebrand factor |
| 135 | S60085S2 | ADMLX=putative adhesion molecule [human,mRNA, 4121 nt, segment 2 of 2] |
| 136 | HSU51334 | Homo sapiens signal transducing adaptor molecule 2A (STAM2) |
| 137 | AF435957 | Homo sapiens Ly-6 antigen/uPA receptor-likedomain-containing protein |
| 138 | NM_000245 | Homo sapiens met proto-oncogene (hepatocytegrowth factor receptor) |
| 139 | XM_044659 | Homo sapiens c-src tyrosine kinase (CSK) |
| 140 | AF061573 | Homo sapiens protocadherin (PCDH8) |
| 141 | HUMMFAP | Homo sapiens extracellular matrix protein (MFAP3)gene |
| 142 | AF081535 | Homo sapiens CDC45L (CDC45L) |
| 143 | HUMCA1XIA | Human alpha-1 type XI collagen (COL11A1) |
| 144 | AB016625 | Homo sapiens OCTN2 gene |
| 145 | AF151899 | Homo sapiens CGI-141 protein |
| 146 | HSU12535 | Human epidermal growth factor receptor kinasesubstrate (Eps8) |
| 147 | HSFCRIB | Human mRNA for high affinity Fc receptor (FcRI) 慴 form’ |
| 148 | HSU74628 | Homo sapiens cell division control related protein (hCDCrel-1) |
| 149 | AF039564 | Homo sapiens retinoblastoma binding protein (RBBP9) |
| 150 | HUMGAPA | Human GTPase-activating protein ras p21 (RASA) |
| 151 | HUMSTK2A | Human protein serine/threonine kinase stk2 |
| 152 | AF144700 | Homo sapiens small zinc finger-like protein (TIM13) |
| 153 | HUMTUBAK | human alpha-tubulin |
| 154 | HUMADCY | Homo sapiens adenyl cyclase-associated protein (CAP) |
| 155 | HSU89329 | Human alternatively spliced microtubule-associatedprotein 2C (MAP2) |
| 156 | BC00051 | Homo sapiens, Insulin-like growth factor 2 |
| 157 | HSU89329 | Human alternatively spliced microtubule-associated protein 2C (MAP2) |
| 158 | BC00051 | Homo sapiens, insulin-like growth factor 2 |
| 159 | AB000529 | Homo sapiens, prostate differentiation factor |
| 160 | HSMAP01 | Human microtuble-associated protein-2 (MAP-2) gene, exon 1 |
| 161 | X67951 | Human mRNA for proliferation-associated gene (pag) |
| 162 | M94250 | Human retinoic acid inducible factor (MK) gene |
| 163 | XM_046278 | Homo sapiens core promoter element binding protein (COPEB) |
| 164 | HSBM40 | Human mRNA for extracellular matrix protein BM-40 |
| 165 | HSU76381 | Homo sapiens fibroblast growth factor (FGF-12b) |
| 166 | HSCALM2S04 | Homo sapiens calmodulin (CALM2) gene, exons 3-6 |
| 167 | HUMID2X | Human helix-loop-helix protein(ID-2) |
| 168 | U20758 | Human osteopontin gene |
| 169 | AF152307 | Homo sapiens protocadherin alpha 11(PCDH-alpha11) |
| 170 | HUMAAMP1X | Homo sapiens angio-associated migratory cell protein (AAMP) |
| 171 | HUMMXI1A | Human MXI1 |
| 172 | AF143536 | Homo sapiens colon cancer-associated protein Mic1(MIC1) |
| 173 | HSU70322 | Human transportin (TRN) |
| 174 | HUMMNMP | Human major nuclear matrix protein |
| 175 | AF071057 | mRNA differentially expressed in GC7901 and GES-1 |
| 176 | AF219140 | Homo sapiens gastric cancer-related protein GCYS-20 |
| 177 | HSU40282 | Homo sapiens integrin-linked kinase (ILK) |
| 178 | HSCA2VR | Human mRNA for pro-alpha 2 (V) collagen chain |
| 179 | HUMPECAM27 | Homo sapiens platelet/endothelial cell adhesion molecule-1 (PECAM-1) gene |
| 180 | XM_165823 | Homo sapiens tumor necrosis factor (TNF superfamily, member 2) (TNF) |
| 181 | D21063 | Homo sapiens MCM2 minichromosome maintenance deficient 2, mitotin |
| 182 | XM_168045 | Homo sapiens CD24 antigen (small cell lung carcinoma cluster 4 antigen) (CD24) |
| 183 | XM_030326 | Homo sapiens CD44 antigen (CD44) |
| 184 | XM_034862 | Homo sapiens interferon regulatory factor 1 (IRF1) |
| 185 | AB025106 | Homo sapiens mRNA for E-cadherin |
| 186 | U73704 | Homo sapiens 48 ku FKBP-associated protein FAP48 |
| 187 | AF380298 | Oncorhynchus mykiss interferon regulatory factor 1 gene, promoter region and partial sequence |
| 188 | L24203 | Homo sapiens ataxia-telangiectasia group D-associated protein |
| 189 | D45906 | Homo sapiens mRNA for LIMK-2 |
| 190 | D21255 | Human mRNA for OB-cadherin-2 |
| 191 | X54925 | Homo sapiens mRNA for type I interstitial collagenase |
| 192 | X05232 | Human mRNA for stromelysin, matrix metalloproteinase 3 |
| 193 | M22612 | Human pancreatic trypsin 1 (TRY1) |
| 194 | HUMGOS8PPC | Human helix-loop-helix basic phosphoprotein (GOS8) |
| 195 | HUMTHBS3 | Homo sapiens thrombospondin 3 (THBS3) gene |
| 196 | HSVECAD | Homo sapiens VE-cadherin |
| 197 | HSBTRCP | Homo sapiens mRNA for beta-transducin repeat containing protein |
| 198 | HUMPROFII | Human profilin II |
| 199 | HSCALT | Homo sapiens mRNA for caltractin |
| 200 | AF091214 | Homo sapiens WRN (WRN) |
| 201 | AF070561 | Homo sapiens clone 24703 beta-tubulin |
| 202 | HUMCD14MCA | Human monocyte antigen CD14 (CD14) |
| 203 | HUMCAM1V | Human vascular cell adhesion molecule 1 |
| 204 | AF032900 | Homo sapiens timing protein CLK-1 |
| 205 | AF070561 | Homo sapiens clone 24703 beta-tubulin |
| 206 | HSUPUE | Homo sapiens mRNA for unknown protein of uterine endometrium |
| 207 | HUMIL8RB | Homo sapiens interleukin 8 receptor beta (IL8RB) |
| 208 | HSERK3 | Homo sapiens ERK3 |
| 209 | AF208045 | Homo sapiens breast cancer-associated antigen BRCAA1 (BRCAA1) |
| 210 | AF081259 | Homo sapiens testis-specific chromodomain Y-like protein (CDYL) |
| 211 | AB022918 | Homo sapiens mRNA for alpha2,3-sialyltransferaseST3Gal VI |
| 212 | AF057036 | Homo sapiens acetylcholinesterase collagen-like tail subunit (COLQ) |
| 213 | AF152497 | Homo sapiens protocadherin beta 4 (PCDH-beta4) |
| 214 | M86752 | Stress-induced phosphoprotein 1 |
| 215 | L04270 | TNF C receptor |
| 216 | AF009674 | Axin 1 |
| Highly expressed genes in precancerous lesions | ||
| 1 | HSU72621 | Human LOT1 |
| 2 | HUMNMOR | Human NAD(P)H: menadione oxidoreductase |
| 3 | AF009227 | Homo sapiens gamma-heregulin |
| 4 | HSU44839 | Human putative ubiquitin C-terminal hydrolase(UHX1) |
| 5 | HUMAAE | Homo sapiens dbpB-like protein |
| 6 | HSU08316 | Human insulin-stimulated protein kinase 1 (ISPK-1) |
| 7 | HUMCD3621 | Human antigen CD36 (clone 21) |
| 8 | Z11899 | Homo sapiens OTF3 mRNA for encoding octamer binding protein 3B |
| 9 | XM_003226 | Homo sapiens vasoactive intestinal peptide receptor 1 (VIPR1) |
| 10 | HUMPAI2B | Human plasminogen activator inhibitor 2 (PAI-2) |
| 11 | HUMACTIIA | Human activin type II receptor |
| 12 | M93718 | Human nitric oxide synthase |
| 13 | HSU46837 | Human RNA polymerase II holoenzyme componentSRB7 (SRB7) |
| 14 | HUMPCNA | Human proliferating cell nuclear antigen (PCNA) gene |
| 15 | HSPCAR | Human mRNA for calcium dependent protease(small subunit) |
| 16 | HSU12140 | Human tyrosine kinase receptor p145TRK-B(TRK-B) |
| 17 | HSTOP2A10 | Homo sapiens topoisomerase II alpha (TOP2A) gene,exons 34 and 35 |
| 18 | HUMYWXD703 | Homo sapiens ADP/ATP carrier protein (ANT-2)gene |
| 19 | HUMKGF | Human keratinocyte growth factor |
| 20 | HS40KDAP | Homo sapiens 40 ku protein kinase related to ratERK2 |
| 21 | HUMPAFAA | Human mRNA for platelet activating factoracetylhydrolase IB gamma-subunit |
| 22 | HUMLPL | Human lipoprotein lipase |
| 23 | HUMMYLCC | Human smooth muscle myosin alkali light chain (MLC 1sm) |
| 24 | HSU10564 | Human CDK tyrosine 15-kinase WEE1Hu (Wee1Hu) |
| 25 | AF022655 | Homo sapiens cep250 centrosome associated protein |
| 26 | D49737 | Homo sapiens mRNA for cytochrome b large subunitof complex II |
| 27 | HUMCD53GLY | Human CD53 glycoprotein |
| 28 | L02867 | Homo sapiens 62 ku paraneoplastic antigen |
| 29 | HUMCALBETB | Human voltage-dependent calcium channel beta-1subunit |
| 30 | HUMEPSURAN | Human surface antigen |
| 31 | AB020647 | Homo sapiens mRNA for KIAA0840 protein |
| 32 | HSU88966 | Human protein rapamycin associated protein(FRAP2) gene |
| 33 | HUMHGLUT1 | Human mRNA for glutamate transporter |
| 34 | U70663 | Human zinc finger transcription factor hEZF(EZF) |
| 35 | HSPTS1R | Homo sapiens mRNA for peroxisomal targetingsignal 1 (SKL type) receptor |
| 36 | HSU61276 | Human transmembrane protein Jagged 1 (HJ1) |
| 37 | HUMMYONM | Human nonmuscle myosin heavy chain (NMHC) |
| 38 | AF016270 | Homo sapiens thyroid hormone receptor coactivating protein |
| 39 | HSU66243 | Human p38 gamma MAP Kinase |
| 40 | HSU41766 | Human metalloprotease/disintegrin/cysteine-richprotein precursor (MDC9) |
| 41 | HUMELF2 | Human translational initiation factor 2 beta subunit(elF-2-beta) |
| 42 | HUMCYCAA | Human somatic cytochrome c (HCS) gene |
| 43 | NM_013217 | Homo sapiens gene for AF-6 |
| 44 | AB017642 | Homo sapiens mRNA for oxidative-stress responsive 1 |
| 45 | AF110956 | Homo sapiens SUMO-1 activating enzyme subunit 1 |
| (SAE1) | ||
| 46 | HUMALR | Human aldehyde reductase |
| 47 | HUMATPSAS | Human gene for ATP synthase alpha subunit (exon1 to 12) |
| 48 | AF052497 | Homo sapiens clone B18 |
| 49 | AB000889 | Homo sapiens mRNA for phosphatidic acidphosphatase 2b |
| 50 | HUMTPARN | Homo sapiens mRNA for tissue plasminogen activator. |
| 51 | AF006082 | Homo sapiens actin-related protein Arp2 (ARP2) |
| 52 | HSU21090 | Human DNA polymerase delta small subunit |
| 53 | HUMVENHK1 | Human voltage-gated potassium channel (HK1) |
| 54 | HUMVTNR | Human cell adhesion protein (vitronectin) receptoralpha subunit |
| 55 | AF091242 | Homo sapiens ATP sulfurylase/APS kinase 2 |
| 56 | HUMIGFBP1 | Human insulin-like growth factor binding protein-1 (IGFBP1) gene |
| 57 | AF047439 | Homo sapiens unknown |
| 58 | AF117386 | Homo sapiens ubiquitin-specific protease (UBP) |
| 59 | AF092129 | Homo sapiens guanine nucleotide binding protein gamma-3 subunit |
| 60 | HUMCOXIV | Human cytochrome c oxidase COX subunit IV(COX IV) |
| 61 | J05412 | Human regenerating protein (reg) gene |
| 62 | AF054162 | Gccys-1, mRNA differentially expressed betweenGC7901 and GES-1 |
| 63 | AF054163 | Gccys-2, mRNA differentially expressed betweenGC7901 and GES-1 |
| 64 | AF054164 | Gccys-3,mRNA differentially expressed betweenGC7901 and GES-1 |
| 65 | AF054165 | Gccys-4, mRNA differentially expressed betweenGC7901 and GES-1 |
| 66 | AF054166 | Gccys-5, mRNA differentially expressed betweenGC7901 and GES-1 |
| 67 | AF054167 | Gccys-6, mRNA differentially expressed betweenGC7901 and GES-1 |
| 68 | NM_003542 | Homo sapiens H4 histone family, member G(H4FG) |
| 69 | XM_032781 | Homo sapiens tubulin, gamma 1 (TUBG1) |
| 70 | XM_083852 | Homo sapiens ribonucleotide reductase M1polypeptide(RRM1) |
| 71 | HSU51586 | Human siah binding protein 1 (siahBP1) |
| 72 | X55181 | Human ETS2 gene |
| 73 | NM_004526 | Homo sapiens MCM2 minichromosomemaintenance deficient 2, mitotin (MCM2) |
| 74 | XM_040900 | Homo sapiens MAP/microtubule affinity-regulatingkinase 3 (MARK3) |
| 75 | XM_083852 | Homo sapiens ribonucleotide reductase M1polypeptide(RRM1) |
| 76 | NM_012145 | Homo sapiens deoxythymidylate kinase(thymidylate kinase) (DTYMK) |
| 77 | X59543 | Ribonucleotide reductase M1 polypeptide |
| 78 | M74542 | Human aldehyde dehydrogenase type III (ALDHIII) |
| 79 | M61855 | Human cytochrome P4502C9 (CYP2C9) |
| 80 | S37730 | Homo sapiens insulin-like growth factor bindingprotein-2 |
| 81 | AB015982 | Homo sapiens EPK2 mRNA for serine/threoninekinase |
| 82 | X67951 | Human mRNA for proliferation-associated gene(pag) |
| 83 | AF127506 | Homo sapiens adenomatosis polyposis coli tumorsuppressor (APC) gene |
| 84 | HT880 | Human Gastric mucin 6 |
| 85 | M63154 | Gastric intrinsic factor |
| Highly expressed genes in normal gastric mucous | ||
| 1 | X05997 | Human mRNA for gastric Lipase |
| 2 | U75272 | Human gastricsin |
| 3 | M63154 | Human intrinsic factor |
| 4 | AF043909 | Homo sapiens gastric mucin (MUC5AC) |
| 5 | L07518 | Homo sapiens mucin |
| 6 | M61853 | Human cytochrome p4502C18 (CYP2C18) |
| 7 | M10942 | Human metallothionein-Ie gene (hMT-Ie) |
| 8 | L15533 | Homo sapiens pancreatitis-associated protein (PAP)gene |
| 9 | Z49107 | Homo sapiens galectin |
| 10 | U52191 | Human SMCY (H-Y) |
| 11 | NM_005522 | Homo sapiens homeo box A1 (HOXA1) |
| 12 | M57732 | Human hepatic nuclear factor 1 (TCF1) |
| 13 | X59770 | Homo sapiens IL-1R2 mRNA for type II interleukin-1 receptor |
| 14 | X76223 | Homo sapiens MAL gene exon 4 |
| 15 | U05259 | Human MB-1 gene |
| 16 | XM_052013 | Homo sapiens polymeric immunoglobulin receptor(PIGR) |
| 17 | U90065 | Human potassium channel KCNO1 |
| 18 | M55422 | Human Krueppel-related zinc finger protein (H-plk) |
| 19 | S78825 | Id1, transcription regulator helix-loop-helix protein |
| 20 | U19948 | Human protein disulfide isomerase (PDIp) |
| 21 | U43522 | Human cell adhesion kinase beta (CAKbeta) |
| 22 | U12139 | Human alphal (XI) collagen (COL11A1) gene, 5region and exon 1 |
| 23 | M14539 | Human factor XIII subunit |
| 24 | X65614 | Homo sapiens mRNA for calcium-binding proteinS100P |
| 25 | AF000560 | Homo sapiens TTF-I interacting peptide 20 |
| 26 | AF002224 | Homo sapiens Angelman Syndrome Gene, E6-APubiquitin protein ligase 3A |
| 27 | U57096 | Human janus kinase 3 (Jak3) |
| 28 | U42600 | Human calcium-activated potassium channel betasubunit |
| 29 | NM_017406 | cAMP responsive element binding protein-like 1 |
| 30 | U04806 | Human FLT3/FLK2 ligand |
| 31 | D84361 | Human p52 and p64 isoforms of N-Shc |
| 32 | Z30425 | Homo sapiens orphan nuclear hormone receptor |
| 33 | M16364 | Human creatine kinase-B |
| 34 | X96924 | Homo sapiens encoding mitochondrial citratetransport protein |
| 35 | HSNM23H1 | Homo sapiens nm23H1 gene |
| 36 | NM_014792 | Homo sapiens KIAA0125 gene product (KIAA0125) |
| 37 | M34041 | Human alpha-2-adrenergic receptor (aipha-2 c2) gene |
| 38 | XM_002444 | Homo sapiens serine threonine kinase 39 (Stk39) |
| 39 | NM_001690 | Homo sapiens ATPase, H+ transporting, lysosomal70 ku, V1 subunitA |
| 40 | L12398 | Human sapiens dopamine receptor D4 (DRD4) |
| 41 | L76465 | Homo sapiens NAD+ dependent 15hydroxyprostaglandin dehydrogenase (PGDH) |
| 42 | U57094 | Human small GTP-binding protein |
| 43 | Z14978 | Homo sapiens mRNA for actin-related protein |
| 44 | X53961 | Human lactotransferrin |
| 45 | M62628 | Human alpha-1 Ig germline C-region membrane-coding region |
| 46 | M84526 | Human adipsin/complement factor D |
| 47 | X04391 | Human lymphocyte glycoprotein T1/Leu-1 |
| 48 | X044533 | Homo sapiens sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) andshort cytoplasmic domain, (semaphoring) 4B(SEMA4B) |
| 49 | AF071054 | Gcys-11, mRNA differentially expressed in cell linesGC7901 and GES-1 |
| 50 | AF063015 | Homo sapiens cell division protein |
| 51 | AF071056 | Gcys-17, mRNA differentially expressed in cell lines GC7901 and GES-1 |
| 52 | AF071058 | Gcys-15, mRNA differentially expressed in cell lines GC7901 and GES-1 |
| 53 | NM_001730 | Homo sapiens Kruppel-like factor 5 (intestinal)(KLF5), mRNA |
| 54 | AB047278 | Arabidopsis thaliana AtNdr 1 mRNA for Ndr kinase |
| 55 | XM_061005 | Homo sapiens similar to Mucin 2 precursor(Intestinal mucin2) |
| 56 | HUM20D9 | Human gene for 2-oxoglutarate dehydrogenase |
| 57 | HSCDC2 | Human CDC2 gene involved in cell cycle control |
| 58 | AF202051 | Homo sapiens NM23-H8 (NME8) |
| 59 | NM_005423 | Homo sapiens trefoil factor 2 (spasmolytic protein 1) (TFF2) |
| 60 | D50419 | Homo sapiens OTK18 |
| 61 | HSU88870 | Human cell division control-related protein 2b(hcdcrel2b) |
| 62 | NM_031942 | Homo sapiens cell division cycle associated 7(CDCA7) |
| 63 | HSU09716 | Human mannose-specific lectin (MR60) |
| 64 | HSU14394 | Human tissue inhibitor of metalloproteinases-3 |
| 65 | Z48314 | Apomucin |
| 66 | M63154 | Gastric intrinsic factor |
| 67 | J05412 | Regenerating protein |
| 68 | M57732 | Hepatic nuclear factor 1 |
| 69 | U70663 | Kruppel-like factor 4 |
| 70 | AB002559 | Syntaxin binding protein 2 |
| 71 | U80226 | GABA transaminase |
| 72 | U05259 | CD79A |
| 73 | X04391 | CD5 |
| 74 | U60800 | CD100 |
| 75 | M74542 | Aldehyde dehydrogenase 3 |
| 76 | X66839 | Carbonic anhydrase IX |
| 77 | L00972 | Cystathionine-beta-synthase |
| 78 | L41688 | UDP-galactose-4 epimerase |
| 79 | J03915 | Chromogranin A |
| 80 | S76942 | Dopamine receptor D4 |
| 81 | D14695 | Herp |
| 82 | D50915 | D50915 |
| 83 | D86961 | HMGIC fusion partner-like 2 |
| 84 | X96924 | Mitochondrial citrate transporter |
| 85 | M16364 | Creatine kinase, brain |
| 86 | M14539 | Factor XIII precursor |
| 87 | U19948 | Protein disulfide isomerase |
| 88 | X65614 | S100 calcium binding protein P |
| Highly expressed genes associated with metastasis | ||
| 1 | NM_004994 | Homo sapiens matrix metalloproteinase 9(gelatinase B, 92 ku type IV collagenase) (MMP9) |
| 2 | XM_053256 | Homo sapiens mucin 1, transmembrane (MUC1) |
| 3 | XM_010702 | Homo sapiens cathepsin K (pycnodysostosis) (CTSK) |
| 4 | NM_002628 | Homo sapiens profiling 2 (PFN2), transcript variant 2 |
| 5 | NM_002128 | Homo sapiens high-mobility group protein 1 (HMG1) |
| 6 | M28130 | Human interleukin 8 (IL8) gene |
| 7 | S3488 | Metastasis-associated gene (human, highlymetastatic lung cell subline Anip) |
| 8 | NM_005231 | Homo sapiens ems1 sequence, transcript variant 1 |
| 9 | XM_059020 | Homo sapiens similar to GPI-anchored metastasis-associated protein homolog |
| 10 | NP_571483 | Vascular endothelial growth factor (VEGF) |
| 11 | I56986 | OPN-a-human (fragment) |
| 12 | AAG31602 | CD44 isoform v3-v6 |
| 13 | AF018733 | 92 ku type IV collagenase precursor (matrixmetalloproteinase-9) (MMP-9) |
| 14 | AF00196 | Octamer-binding transcription factor 2 (OTF-2) |
| 15 | XM_055254 | Homo sapiens fibronectin 1 (FN1) |
| Risk factor genes | ||
| 1 | V01555 | Epstein-Barr virus (EBV) genome, strain B95-8 |
| 2 | AF275307 | H pylori plasmid pHPM8 (cagA) |
| 3 | AF275307 | H pylori plasmid vacA |
| 4 | AF275307 | H pylori plasmid Urase |
| 5 | AF431736 | Human herpesvirus 1 strain KOS ICPO gene |
| 6 | Z86099 | Herpes simplex virus type 2 (strain HG52) |
| 7 | AF477385 | Human papillomavirus type 16 E7 gene |
| 8 | AX742207 | Human hepatitis virus 11 type |
The gene expression profiles of each specimen obtained by biochip were stored together with patient clinical data including follow-up treatments until death. The data files were incorporated into a computer database by CGO software, including patients’ disease history and all screened results such as name, file number, sex, age, address, telephone, e-mail address, marital status, blood type, body mass, disease history, imaging examination, pathological examination, serum examination, blood examination, cytogenetic report, gene array report. The prewarning data were added with new content. These data would be available on Gastric Cancer Information Web presided by Dr. Cui at http://www.37c.com.cn.
A total of 412 genes were selected as the main diagnostic genes, including 216 genes that displayed higher expression levels in cancer tissues than in non-cancer tissues, 85 genes with higher expression levels in precancerous lesions than in cancer tissues and 88 genes that exhibited higher expression levels in normal tissues than in gastric cancer tissues and pre-cancerous tissues. We selected 15 genes associated with metastasis of gastric cancer as metastasis biomarkers, 8 risk factor genes as reference biomarkers to predict the development of pre-cancerous lesions (Table 1). The critical threshold values to distinguish normal gastric mucosa from pre-cancerous lesion and gastric cancer were decided and were summarized in Table 2.
| Gene classification | Gastric cancer tissue (GC/N) | Precancerous lesion (PC/N) | Normal gastric mucosa (N*/GC or N*/PC) |
| 216 genes associated with gastric cancer | 6.01±2.40 | 1.18 ±0.47 | < 0.75 |
| 85 genes associated with precancerous lesions | 1.32±0.53 | 4.86±1.94 | 2.54±0.41 |
| 88 genes associated with normal mucosa | 1.31±0.54 | 2.50±0.75 | 5.42±2.17 |
| 15 genes associated with metastasis of gastric cancer | 5.81±2.32 (M)2.32±1.19 (N1) | 1.13±0.58 | 0.65±0.35 |
| 8 genes associated with risk factors | >2.0 |
All 412 genes and critical threshold values to distinguish normal gastric mucosa from precancerous lesion and gastric cancer were compiled into an analysis software, which can automatically provide analysis reports by analyzing the provided microarray test results. The analysis software for examination results of prewarning system of gastric cancer locates on the website http://shasta.mpi-stuttgart.mpg.de/array/form.html. The software cannot be downloaded until it is confirmed to be very effective and complete.
Two new biomarkers brcaa1 and ndr1 (NM_007271) were identified. Brcaa1 (AF208045) showed no or low-expression levels in normal gastric mucosa and high-expression level in gastric cancer There was a statistically significant difference in expression levels between normal gastric mucous tissues and gastric cancer tissues (P<0.01, Figure 3), indicating that higher expression of brcaa1 was closely associated with gastric cancer stage. Further analysis indicated that higher expression of brcaa1 appeared to have no correlation with pathological types of gastric cancer (P>0.05, data not shown). Conversely, ndr1 (NM_007271) displayed higher expression levels in normal gastric tissues and no or lower expression in gastric cancer, and there was a statistically significant difference in expression levels between normal gastric mucous tissues and gastric cancer tissues (P<0.01), indicating that higher expression level of ndr1 was closely associated with normal stage of gastric mucosa tissues.
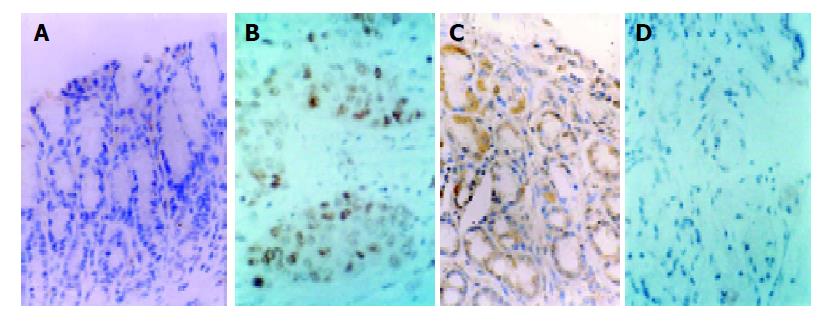
Brcaa1 protein exhibited higher expression in 60 gastric cancer tissues, lower or no expression in 30 normal gastric mucosa tissues. There was a statistically significant difference in expression levels between gastric cancer tissues and normal gastric mucous tissues (P<0.01, Figure 4A). The result indicated that higher expression of brcaa1 was associated with gastric cancer stage. Ndr1 protein exhibited higher expression in 30 normal gastric mucosa tissues, lower or no expression in 60 gastric cancer tissues. There was a statistically significant difference in expression levels between normal gastric mucous tissues and gastric cancer tissues (P<0.01, Figure 4B). The result indicated that higher expression of ndr1 was closely associated with normal stage of gastric mucous tissues.
The development of normal gastric mucosa into gastric cancer is a complex process. Previous research in the pathology of gastric cancer demonstrated that normal gastric mucosa could gradually develop into pre-cancerous lesions under special conditions, eventually evolving toward gastric carcinoma. During the periods from normal gastric mucosa to gastric cancer, it has not been shown how many genes are involved at different stages of cancer development. The cDNA microarray technology could provide an efficient tool to address the difficulties in screening and quantifying expression levels of a large number of genes[7-10]. So far there are some reports associated with gene expression profiles of gastric cancer based on biochip[11,12]. However, the problem of early gastric cancer detection is still not solved satisfactorily. In the present study, we tried to establish a prewarning system of gastric cancer based on biochip and CAD technique to solve the problem of early gastric cancer detection.
Firstly, two high-density microarrays with 8 464 human cDNA sites were used to screen two pairs of gastric cancer tissues and 389 genes associated with three stages of gastric cancer development such as normal gastric mucosa, precancerous lesion and gastric cancer were obtained. The selected 389 genes were used as main diagnostic genes on the prewarning chip, 15 genes associated with metastasis of gastric cancer as diagnostic genes of metastasis stages, 8 risk factor genes as reference biomarkers to predict the development of precancerous lesions.
A total of 412 genes were selected to fabricate the low-density chip, which was used to screen 150 clinical specimens. It was found that the gene expression levels in normal, pre-cancerous lesion and cancer tissues were significantly different as expected. CAD software and statistical methods were used to identify key genes and their critical threshold values characterizing different tissue status. Two hundred and sixteen genes displayed higher expression levels in cancer tissues than in non-cancer tissues, 85 genes exhibited higher expression levels in precancerous lesions than in cancer tissues, and 88 genes exhibited higher expression levels in normal tissues than in gastric cancer and precancerous tissues (Table 1). The critical threshold values to distinguish normal gastric mucosa from precancerous lesion and gastric cancer were identified (Table 2). With the above-mentioned standards, the 150 specimens could be clearly grouped according to their tissue status determined in pathology diagnosis. Therefore, we considered that the established standard had a great potential in the detection of early gastric cancer. Based on these selected genes and critical threshold values characterizing three stages of gastric cancer development, an analysis software was developed which could analyze the examination results of 412 genes achieved by biochip and provide automatically an analysis report. The software remained to be optimized. These expression profiles obtained from all these specimens and available clinical data had been compiled into a prewarning data library of gastric cancer by CGO software, and these detailed data would be very useful for the further research and therapy of gastric cancer.
From Table 2, it appeared reasonable to define integrate markers of GC, PC, NU consisting of many genes, instead of individual genes, to distinguish three kinds of gastric tissues status. Once gastric cancer was diagnosed, the expression levels of 15 metastasis genes could be subjected to focal studies to identify whether the cancer metastasized, and to speculate the prognosis of the cancer patients. These results could also be complemented with supporting evidence from patient’s disease history, for example, discomfort or pain in the gastric area, body mass loss in a short time, etc. If a precancerous lesion was diagnosed, the expression levels of risk factor genes might be analyzed as indicators on how fast such lesion would lead to cancer[13]. One may also establish and search the prewarning database library to compare similar patients to make a best treatment plan. The diagnosis and treatment information associated with gastric cancer can also be obtained from gastric cancer information web presided over by Dr. Cui http://www.37c.com.cn. The prewarning database of gastric cancer is available on gastric cancer information web. The analysis software of examination results of the prewarning system of gastric cancer locates on the website http://shasta.mpi-stuttgart.mpg.de/array/form.html.
Two new biomarkers have been identified of diagnostic value, brcaa1 (AF208045)[14] and ndr1 (NM_007271). Brcaa1 showed no or low-expression levels in normal gastric mucosa and high-expression level in gastric cancer, and appeared to have no correlation with pathological types of gastric cancer. Conversely, ndr1 displayed high-expression levels in normal gastric tissues and no or lower expression in gastric cancer. These results were also confirmed by Northern blot and immunohistochemistry analysis. These two biomarkers may be very useful for distinguishing benign from malignant gastric mucosa lesions.
Gastric cancer specimens from different patients were found to display some variability in gene expression profiles. The reasons could be attributed to variations in specimens, lesion types and the number of cells collected. Moreover, variations among individuals may pose a serious challenge to diagnosis accuracy. In cases of doubt, it would be advisable to analyze microarray results together with clinical symptoms of patients and pathological results. It is very difficult to devise gene expression profiles to further classify the specimens consistent with pathology types such as atrophic gastritis, intestinal gland metaplasia, atypical hyperplasia, etc. Of course, new methods of disease classification can be defined according to gene expression profiles and DNA levels (mutation, deletion and amplification). Such methods may not be fully consistent with pathology classification, but nevertheless may be appropriate for future clinical applications. In the near future, pathological diagnosis will remain a useful and complementary diagnostic tool.
To test the generality of this standard, we collected randomly some autopsy specimens and screened them with fabricated gastric microarrays. Simultaneously, pathology diagnosis was performed on the same specimens. We found that the results achieved by the microarray were highly identical with traditional pathological results. In another paper, we have reported these results in detail[15,16].
In summary, further studies will lead to a more complete prewarning database library. The prewarning database, together with miniaturized microarray techniques, will be used to further improve the accuracy and reliability of the prewarning system for gastric cancer[16].
The authors thank Professor Deng-Cheng Li of Xi’an Jiaotong University for his CAD software.
Edited by Wang XL and Gabbe M
| 1. | van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6904] [Cited by in RCA: 6356] [Article Influence: 276.3] [Reference Citation Analysis (0)] |
| 2. | Cui DX, Yan XJ, Wang F, Zhao JR, Su CZ. Studies on differentially expressed genes of gastric cancer by mRNA differential display. Shenywu Huaxue Yu Shengwu Wuli Jinzhan. 2000;27:379 382. |
| 3. | Cui DX, Yan XJ, Wang F, Su CZ. New strategy of cloning of differentially expressed genes. Shenywu Huaxue Yu Shengwu Wuli Jinzhan. 2000;27:362 364. |
| 4. | Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233-240. [PubMed] |
| 5. | Inoue H, Matsuyama A, Mimori K, Ueo H, Mori M. Prognostic score of gastric cancer determined by cDNA microarray. Clin Cancer Res. 2002;8:3475-3479. [PubMed] |
| 6. | Sipponen P. Gastric cancer: pathogenesis, risks, and prevention. J Gastroenterol. 2002;37 Suppl 13:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1935] [Cited by in RCA: 1253] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 8. | Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012-7017. [PubMed] |
| 9. | Bumm K, Zheng M, Bailey C, Zhan F, Chiriva-Internati M, Eddlemon P, Terry J, Barlogie B, Shaughnessy JD. CGO: utilizing and integrating gene expression microarray data in clinical research and data management. Bioinformatics. 2002;18:327-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Sugiyama T, Hige S, Asaka M. Development of an H. pylori-infected animal model and gastric cancer: recent progress and issues. J Gastroenterol. 2002;37 Suppl 13:6-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | Lee JH, Koh JT, Shin BA, Ahn KY, Roh JH, Kim YJ, Kim KK. Comparative study of angiostatic and anti-invasive gene expressions as prognostic factors in gastric cancer. Int J Oncol. 2001;18:355-361. [PubMed] |
| 12. | Sepulveda AR, Tao H, Carloni E, Sepulveda J, Graham DY, Peterson LE. Screening of gene expression profiles in gastric epithelial cells induced by Helicobacter pylori using microarray analysis. Aliment Pharmacol Ther. 2002;16 Suppl 2:145-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Petersson F, Borch K, Franzén LE. Prevalence of subtypes of intestinal metaplasia in the general population and in patients with autoimmune chronic atrophic gastritis. Scand J Gastroenterol. 2002;37:262-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Cui DX, Gao TW, Jin GQ, Sun TB, Sarai A. Cloning and characterization analysis of BRCAA1. identification. Ziran Zazhi. 2003;25:356 358. |
| 15. | Cui DX, Zhang L, Zhang LX, Su CZ, Jin GQ, Xu JR, Yan XJ, Sun TB, Fan DM, Gao HJ. A microarray based prewarning system of gastric cancer. IFMBE Proc. 2002;3:770 773. |
| 16. | Cui D, Gao H. Advance and prospect of bionanomaterials. Biotechnol Prog. 2003;19:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |