Published online Dec 21, 2005. doi: 10.3748/wjg.v11.i47.7405
Revised: July 18, 2005
Accepted: July 20, 2005
Published online: December 21, 2005
AIM: To examine the gene expression profile of gastric cancer (GC) by combination of laser capture microdissection (LCM) and microarray and to correlate the profiling with histological subtypes.
METHODS: Using LCM, pure cancer cells were procured from 45 cancerous tissues. After procurement of about 5 000 cells, total RNA was extracted and the quality of RNA was determined before further amplification and hybridization. One microgram of amplified RNA was converted to cDNA and hybridized to cDNA microarray.
RESULTS: Among 45 cases, only 21 were qualified for their RNAs. A total of 62 arrays were performed. These included 42 arrays for cancer (21 cases with dye-swab duplication) and 20 arrays for non-tumorous cells (10 cases with dye-swab duplication) with universal reference. Analyzed data showed 504 genes were differentially expressed and could distinguish cancerous and non-cancerous groups with more than 99% accuracy. Of the 504 genes, trefoil factors 1, 2, and 3 were in the list and their expression patterns were consistent with previous reports. Immunohistochemical staining of trefoil factor 1 was also consistent with the array data. Analyses of the tumor group with these 504 genes showed that there were 3 subgroups of GC that did not correspond to any current classification system, including Lauren’s classification.
CONCLUSION: By using LCM, linear amplification of RNA, and cDNA microarray, we have identified a panel of genes that have the power to discriminate between GC and non-cancer groups. The new molecular classification and the identified novel genes in gastric carcinogenesis deserve further investigations to elucidate their clinicopathological significance.
- Citation: Wu MS, Lin YS, Chang YT, Shun CT, Lin MT, Lin JT. Gene expression profiling of gastric cancer by microarray combined with laser capture microdissection. World J Gastroenterol 2005; 11(47): 7405-7412
- URL: https://www.wjgnet.com/1007-9327/full/v11/i47/7405.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i47.7405
Gastric cancer (GC) is the second most common cause of cancer-related deaths in the world[1]. In Taiwan, GC ranks the fourth cancer-related death and caused more than 2 000 deaths annually[2]. The prognosis of GC depends mainly on early detection and adequate surgical resection. Although endoscopy with biopsy has been effectively used since 1980s for early diagnosis, the proportion of early to advanced GC being found through this method has not appreciably increased in recent years[3]. In spite of the current surgical techniques and chemotherapy that have made significant improvements, the cure rate for advanced GC remains low and the morbidity remains high[4]. Thus, to improve its detection and therapy, understanding of the pathogenesis and biologic features of GC is crucial.
Gastric carcinogenesis is a multi-factorial and multi-step process accompanied by accumulation of alterations of critical growth regulatory genes[5]. Delineating these genes involved may lead to important new insights into carcinogenesis[6]. Despite the fact that some interesting and promising genetic alterations have been elucidated[7], previous studies of gastric carcinogenesis were incomprehensive and inconclusive. The overall information of the genetic alterations is scanty due to technical problems of analyses. Firstly, the value of even the most sophisticated genetic testing methods will be limited if the inputs of genetic materials are not derived from pure populations or are contaminated by the wrong cells. In solid neoplasm such as GC, stromal and inflammatory cells usually intermingled with cancer cells. Therefore, special procedures to isolate cancer cells from heterogeneous tissues are mandatory. Secondly, most of previous studies on genetic alterations of GC have focused on selected genes or chromosomal regions known in other cancers. These obstacles may be overcome using the recently developed techniques of laser capture microdissection (LCM) and microarray. LCM allows for the rapid, reliable, and accurate procurement of cells from specific microscopic regions of tissue sections under direct visualization[8]. It affords the opportunity to perform molecular genetic analysis of pure populations of malignant cells in their native tissue environment. On the other hand, the advent of high-density cDNA microarray technology with its capacity for simultaneous monitoring of thousands of genes, provides a unique opportunity for high-throughput genetic analysis of cancer[9]. For GC, several investigators have demonstrated the use of DNA microarray is beneficial for elucidation of gastric carcinogenesis[10-24]. However, to our knowledge, combined analyses of LCM and microarray in GC remain scanty[22-24]. Therefore, we aimed to examine gene expression profiles of GC by these two techniques.
A total of 45 cancerous tissues and their respective non-cancerous tissues obtained at operation from patients with GC were collected and immediately frozen in liquid nitrogen. Gastric cancer and normal cells were stained by HistoGene LCM Frozen Section Staining Kit and laser capture microdissected by using a Pix Cell II LCM system (Arturus, USA). Malignant and normal cells were captured in a number of about 5 000 cells and their total RNAs were isolated by using PicoPure RNA extraction kit (Arturus, USA). The quality of RNA was determined by Bioprocessor before further amplification and hybridization.
Linear RNA amplification was performed by using the RiboAmp kit (Arturus, USA). Two rounds of RNA amplification were performed to obtain enough amplified RNA (aRNA) for a microarray experiment. To serve as reference in cDNA microarray comparison, a human reference RNA pooled from 9 cell lines (Stratagene, USA) was amplified identically. cDNA was transcribed from aRNA at a quantity of 1.5 µg per channel in the presence Cy3- or Cy5-dUTP by using Cyscribe First-Strand cDNA Labelling kit (Amersham Biosciences, USA). Free conjugated dUTP was removed by Millipore Microcon YM-30 column. Cy3-labeled cDNA was pooled with Cy5-labeled reference probe in 30 µL of hybridization solution and hybridized to an Agilent human 1 cDNA microarray (Agilent Technology, USA). Hybridization was carried out at 65 °C for 17 h in a humidified dark chamber (Genetix, UK). After hybridization, slides were washed at the following condition: 2 X SSC/0.1 g/L SDS at 60 °C for 10 min, 2X SSC at room temperature for 10 min, and 0.2X SSC at room temperature for 10 min.
Washed microarrays were scanned with a Virtek fluorescence reader (Virtek, CA, USA) at 535 nm for Cy3 and 625 nm for Cy5. Scanned images were analyzed by using Array-Pro image acquisition software (Media Cybernetics, USA), an image analysis algorithm was used to quantify the signal and background intensity for each target element. Data normalization was performed by Iowess method using R package (written by Terry Speeds Microarray Data Analysis Group, University of Berkerley, USA). Measurements from dye-swap replicates were average after normalization. A software developed by researchers in Stanford University-Prediction Analysis of Microarray (PAM) and Spotfire software was utilized to analyze the data.
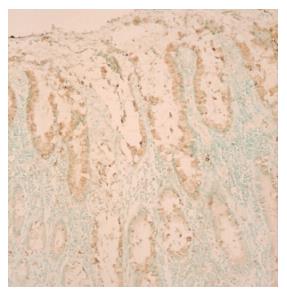
Immunostaining for trefoil factor 3 protein was performed by using a standard avidin-biotin-peroxidase complex detection system[25]. The monoclonal antibody used for this study was purchased from Signet Laboratory (Bedham, MA, USA). In brief, 5-µm sections were dewaxed, microwaved, and rehydrated. Endogenous peroxidase activity and non-specific bindings were blocked by incubation with 30 mL/L hydrogen peroxide (H2O2) and non-immune serum, respectively. The slides were then incubated sequentially at 4 °C with the primary mouse monoclonal antibody overnight, a biotinylated goat anti-mouse secondary antibody for 30 min, peroxidase-conjugated streptavidin for 10 min and finally diaminobenzidine tetrachloride/H2O2 chromogen substrate for 10 min. Slides were then counterstained with Mayer’s hematoxylin. Negative control sections were prepared by substituting the primary antibody with buffered saline. The percentage of positively stained cells was determined for each tumor section as well as its adjacent intestinal metaplasia and non-metaplastic epithelium. The immunostaining for trefoil factor 3 was registered as negative only if less than 5% of the cells showed a positive staining.
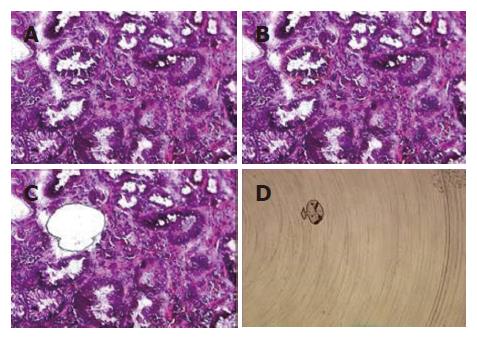
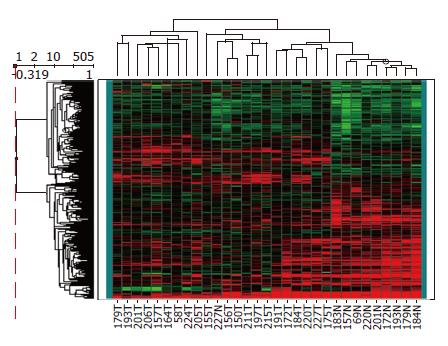
Among the 45 subjects with GC, only 21 (12 intestinal and 9 diffuse subtypes by Lauren classification; 19 advanced and 2 early gastric cancer by depth of invasion) of their respective RNAs were qualified for further analyses after capture of malignant cells by LCM. Figure 1 shows a representative example of LCM. A total of 62 arrays were then performed, including 42 arrays for cancer (21 cases with dye-swap duplication) and 20 arrays for non-tumorous cells (10 cases with day-swap duplication) with universal reference. PAM analyses showed differential expression of 504 genes (Tables 1 and 2) could distinguish the cancerous and non-cancerous groups with more than 99% accuracy. Of the 504 genes, trefoil factors 1, 2, and 3 were in the list and their expression patterns (down-regulation of trefoil factors 1 and 2, and up-regulation of trefoil factor 3 in cancer cases) were consistent with previous reports. In addition, we verified expression status of trefoil factor 3 in GC by immunohistochemical staining (Figure 2). Comparison of the tumor group and non-cancerous group with these 504 genes showed that there were 3 subgroups of GC (Figure 3) that did not correspond to any current classification system, including Lauren’s classification.
| Biosynthesis |
| Argininosuccinate synthetase [BC009243], hydroxymethylbilane synthase [BC000520], hypothetical protein CL640 [BC008804], methylenetetrahydrofolate dehydrogenase (NADP+-dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase [BC001014] |
| Cell adhesion |
| Collagen, type XVIII, alpha 1 [AF01808], mesothelin [U40434], CD9 antigen (p24) (X60111), collagen, type VI, alpha 3 [X52022], thrombospondin 2 [L12350], immunoglobulin superfamily containing leucine-rich repeat [AB003184] |
| Cell death |
| Nerve growth factor receptor (TNFR superfamily, member 16) [M14764], phosphoprotein enriched in astrocytes 15 [AF153274], programmed cell death 5 [AF014955] |
| Cell growth and/or maintenance |
| Cell division cycle 25B [S78187], cyclin D2 [D13639], hippocalcin [BC001777], anillin, actin binding protein (scraps homolog, Drosophila) [AF273437], afamin [L32140], chromosome 14 open reading frame 58 [AK000378], C1q and tumor necrosis factor related protein 1 [AF32984], low density lipoprotein receptor-related protein 8, apolipoprotein e receptor [D86407], enhancer of rudimentary homolog (Drosophila) [U66871], tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) [D11139], insulin receptor substrate 2 [AF073310], low density lipoprotein receptor-related protein 8, apolipoprotein e receptor [D50678], thyroid hormone receptor interactor 10 [AJ000414], collagen, type I, alpha 1 [Z74615], ATPase, Na+/K+ transporting, beta 3 polypeptide [AF005896], T-LAK cell-originated protein kinase [AB027249], platelet-derived growth factor receptor, beta polypeptide [J03278], nucleolar protein 1 120 ku [BC000656], keratin 7 [BC002700], oncostatin M receptor [BC010943], tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) [BC000866], insulin-like growth factor 2 (somatomedin A) [S77035], karyopherin (importin) beta 1 [L38951], nuclear transport factor 2 (NTF-2) (Placental protein 15) (PP15) [X07315], nucleolar protein 1 120ku [X55504], thymosin beta-10 [S54005] |
| Cell motility |
| Hyaluronan-mediated motility receptor (RHAMM) [AF03286], sorcin [M32886], troponin T1, skeletal, slow [BC010963] |
| Cell-cell signaling |
| Ephrin-A2 [U92896], midkine (neurite growth-promoting factor 2) [X55110], prostaglandin I2 (prostacyclin) receptor (IP) [D25418] |
| Immune response |
| Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) [X14831], Human 1-8D gene from interferon-inducible gene family [X57351], GTP binding protein over-expressed in skeletal muscle [U10550], indoleamine-pyrrole 2,3 dioxygenase [M34455], interferon induced transmembrane protein 1 (9-27) [J04164] |
| Macromolecule metabolism |
| Cathepsin L2 [AB001928], PAK2 [AF09213], ribosomal protein, large P2 [M17887], hypothetical protein FLJ22761 [AK026414], KIAA0205 [D86960], proteasome (prosome, macropain) subunit, alpha type, 1 [D00759], protein tyrosine phosphatase type IVA, member 1 [U48296], peptidylprolyl isomerase (cyclophilin)-like 1 [BC003048], ribosomal protein L14 [BC009294], pyruvate kinase, muscle [BC007952], proteasome (prosome, macropain) subunit, alpha type, 7 [BC001895], isocitrate dehydrogenase 3 (NAD+) alpha [U07681], transglutaminase 4 (prostate) [U31905] |
| Organogenesis |
| Secreted protein, acidic, cysteine-rich (osteonectin) [M25743], caudal type homeo box transcription factor 2(CDX2) [AJ278434], transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) [M36711] |
| Regulation of metabolism |
| Homeo box B7 [BC015345], B-cell receptor-associated protein 37 [AF126021], Mediterranean fever [AJ003147], homeo box B7 [M16937], homeo box D4 [X17360], TIP49 [AB012122], sin3-associated polypeptide, 18 ku [AF153608] |
| Response to stress |
| Complement component 2 [X04481], complement component 3 [K02765], inhibin, beta A (activin A, activin AB alpha polypeptide) [X57579], serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 [K02212], serine (or cysteine) proteinase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) [D83174], meiotic recombination (S. cerevisiae) 11 homolog A [AF073362], RecQ protein-like 4 [BC013277] |
| Signal transduction |
| Secreted frizzled-related protein 4 [AF026692], lymphocyte antigen 6 complex, locus E(RIG-E) [Z68179], dimethylarginine dimethylaminohydrolase 2 [BC001435] |
| Unclassified |
| Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 [BI826471], apolipoprotein C-I [AV709433], argininosuccinate synthetase [BF305206], ATPase, Ca++ transporting, plasma membrane 4 [AI885833], ATPase, Na+/K+ transporting, beta 3 polypeptide [AU151263], B-cell associated protein [BG771222], BTG family, member 2 [BG750101], cadherin 3, type 1, P-cadherin (placental) [NM001793], calcium modulating ligand [AW474551], carbonyl reductase 3 [AI658832], cathepsin Z [AI913006], CDC-like kinase 2 [BG286233], cell division cycle 25B [BF976307], CGI-01 protein [AK027886], chloride intracellular channel 1 [AA291390], claudin 4 [BI760179], claudin 7 [AI279608], collagen, type I, alpha 1 [AW577407], collagen, type X, alpha 1 (Schmid metaphyseal chondrodysplasia) [NM000493], core-binding factor, beta subunit [NM001755], D123 gene product [BE792735], deoxythymidylate kinase (thymidylate kinase) [BF312526], diubiquitin [AW270961], DKFZP566C134 protein [BG621382], E2F transcription factor 3 [AU153511], epithelial protein lost in neoplasm alpha [AAF23756], exostoses (multiple) 1 [BF057267], FK506-binding protein 4 (59 ku) [NM002014], follistatin-like 3 (secreted glycoprotein) [NM005860], forkhead box M1 [AL525810], glutamine-fructose-6-phosphate transaminase 2 [AK001242], GTP-binding protein over-expressed in skeletal muscle [AW297828], guanylate cyclase 2C (heat stable enterotoxin receptor) [NM004963], heme oxygenase (decycling) 1 [BI596354], HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2 [BG167159], Homo sapiens cDNA FLJ11796 fis, clone HEMBA1006158, highly similar to Homo sapiens transcription factor forkhead-like 7 [FKHL7] gene [BI914918], homo sapiens mRNA for short form of beta II spectrin, partial [AA131993], human cDNA: FLJ22528 fis, clone HRC12825 [AK026181], human cDNA: FLJ22998 fis, clone KAT11985, highly similar to AB000712 Human hCPE-R mRNA for CPE-receptor [AK026651], human chromosome 20 clone h119, complete sequence [AF312913], human claudin 3 [CLDN3] gene, [AF007189], regenerating islet-derived family, member 4 [AF254415], human genomic DNA, chromosome 22q11.2, clone KB1125A3 [AP000350], likely ortholog of mouse membrane bound C2 domain containing protein [AB018290], KIAA0930 protein [AB02314], hypothetical protein LOC90499 [AL137712], fuse-binding protein-interacting repressor [AF190744], Thy-1 cell surface antigen [M11749], biglycan [BC002416], CK2 interacting protein 1; HQ0024c protein [BC010149], hypothetical protein FLJ12436 [BC011993], tissue inhibitor of metalloprotease 1 (erythroid potentiating activity, collagenase inhibitor) [BC000866], hypothetical protein FLJ20277 [M11749], hypothetical protein FLJ22393 [H45848],hypothetical protein PRO1489 [AI536671], interferon induced transmembrane protein 1 (9-27) [BG506643], interferon induced transmembrane protein 3 (1-8U) [BE886918], interleukin 13 [NM002188], interleukin 8 [BG777366], JM4 protein [BI818455], keratin 7 [BI094014], keratin 7 [AA307373], KIAA0747 protein [BG744628], low density lipoprotein receptor-related protein 8, apolipoprotein e receptor [BF110337], lymphocyte antigen 6 complex, locus E [BM101706], matrix metalloproteinase 7 (matrilysin, uterine) [NM002423], membrane protein of cholingeric synaptic vesicles [BG760616], mesothelin [AI813749], midkine (neurite growth-promoting factor) [AI784469], msh (Drosophila) homeo box homolog 1 [AI912103], MyoD family inhibitor [BG675569], neural precursor cell expressed, developmentally down-regulated 5 [AU131176], neuromedin U [BG661038], NIPSNAP, C. elegans, homolog 1 [BC006473], nucleolar and coiled-body phosphoprotein [AL553791], platelet-derived growth factor receptor, beta polypeptide [BI755841], procollagen-lysine, 2-oxogutarate 5-dioxygenase 3 [AL544817], proliferating cell nuclear antigen [AA523378], proliferation-associated 2G4, 38KD [BI088072], protein C receptor, endothelial (EPCR) [BG831881], pyruvate kinase, muscle [BF690275], rhoG [BG338917], regulatory factor X, 5 [AW027312], ribosomal protein L23 [AI349581], ribosomal protein L35 [BF310946], ribosomal protein L37 [BG032793], ribosomal protein S16 [BG765030], ribosomal protein S3 [AA593872], S100 calcium-binding protein A4 [AV713821], serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin) [BG567810], sigma receptor (SR31747 binding protein 1) [AL571572], small inducible cytokine subfamily A (Cys-Cys), member 19 [W07401], superoxide dismutase 2, mitochondrial [BG773219], survival of motor neuron 1, telomeric [AA029190], thymosin, beta 10 [AV707021], thyroid hormone receptor interactor 10 [N98668], transKetolase [BI195990], translocase of outer mitochondrial membrane 34 [BE798732], trefoil factor 3 (intestinal) [AA633399], tumor suppressing subtransferable candidate 3 [BE670374], zinc finger protein 267 [AU128446] |
| Biosynthesis |
| 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase [D89976], adenosine monophosphate deaminase 1 (isoform M) [M60092], histidine decarboxylase [D16583], Human histidine decarboxylase (HDC) [M60445] |
| Catabolism |
| Human bilirubin UDP-glucuronosyltransferase isozyme 1 [M57899], Human placental cDNA coding for 5’nucleotidase (EC 3.1.3.5) [X55740] |
| Cell adhesion |
| Collagen, type IV, alpha 6 [D21337], collagen, type XVII, alpha 1[U76585], tenascin C (hexabrachion) [X78565], dermatopontin [AL049455], Human pancreatitis associated protein [M84337], sialic acid binding Ig-like lectin 11 [AF337818], Macaque brain cDNA clone:QflA-14173, full insert sequence [AB062939] |
| Cell death |
| Baculoviral IAP repeat-containing 1 [U80017], CD3E antigen, epsilon polypeptide (TiT3 complex) [X03884], programmed cell death 4 (neoplastic transformation inhibitor) [U96628] |
| Cell growth and/or maintenance |
| Core-binding factor, runt domain, alpha subunit 2; translocated to, 1; cyclin D-related [S69002], ATPase, Ca++ transporting, ubiquitous [Z69881], ATP-binding cassette, sub-family C (CFTR/MRP), member 5 [AB005659], B-cell CLL/lymphoma 3 [M31732], CDC7 (cell division cycle 7, S. cerevisiae, homolog)-like 1 [AF015592], gastrokine 1 [AB039886], Human chondroadherin mRNA, complete cds [AF371328], CDC28 protein kinase regulatory subunit 2 [X54942], solute carrier family 39 (zinc transporter), member 6 [U41060], ATPase, H+/K+ exchanging, alpha polypeptide [M63962], golgi autoantigen, golgin subfamily a, 4 [U31906], ATPase, H+/K+ exchanging, beta polypeptide [M75110], solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 [AB037669], transcription factor-like 5 (basic helix-loop-helix) [AF070992], kinesin family member 11 [U37426], cholecystokinin B receptor [D13305], choline kinase alpha [D10704], sodium channel, nonvoltage-gated 1, gamma [X87160], MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) [D28480], sodium channel, nonvoltage-gated 1 alpha [X76180], oxysterol binding protein-like 7 [AF323729], zinc finger protein 145 (Kruppel-like, expressed in promyelocytic leukemia) [AF060568], solute carrier organic anion transporter family, member 2A1 [U70867], aurora kinase B [AF004022], CDC-like kinase 1 [M59287], trefoil factor 1 [X52003], quiescin (Q6) [U97276], signal recognition particle receptor (‘docking protein’), clone MGC:1609 IMAGE:3528505, mRNA, complete cds [BC001162], villin-like, clone MGC:656 IMAGE:3356294, mRNA, complete cds. [BC000243], inhibitor of DNA binding 1, dominant negative helix-loop-helix protein [S78825], insulin-like growth factor binding protein 2 36 ku [S37730], Macaque somatostatin I mRNA, complete cds. solute carrier family 9 (sodium/hydrogen exchanger), isoform 1 (antiporter, Na+/H+, amiloride sensitive) [S68616], platelet-derived growth factor receptor, alpha polypeptide [M21574], SH3-domain GRB2-like 2 [AF036268], solute carrier family 9 (sodium/hydrogen exchanger), isoform 1 (antiporter, Na+/H+, amiloride sensitive) [M96067], thiosulfate sulfurtransferase (rhodanese) [BI820468], thrombomodulin [M16552] |
| Cell-cell signaling |
| NAD(P)H dehydrogenase, quinone 1 [J03934], dopamine receptor D5 [BC009748], monoamine oxidase A [M69226] |
| Digestion |
| Japanese Macaque (Macaca fuscata) mRNA for pepsinogen A-2/3. [X59755] |
| Electron transport |
| Cytochrome P450, family 4, subfamily F, polypeptide 12 [AY008841], cytochrome P450, family 2, subfamily C, polypeptide 18 [M61853], cytochrome P450, family 4, subfamily F, polypeptide 11 [AF23608], cytochrome P450, family 2, subfamily S, polypeptide 1 [AF33527], dual oxidase 1 [AF21346], cytochrome c oxidase subunit Vic [BC000187] |
| Immune response |
| Diacylglycerol kinase, delta 130 ku [D63479], Fc fragment of IgG, high affinity Ia, receptor for (CD64) [X14356], beta-site APP-cleaving enzyme 2 [AL163285], X-box binding protein 1 [L13850] |
| Macromolecule metabolism |
| Sphingomyelin phosphodiesterase, acid-like 3A [Y08136], calpain 9 (nCL-4) [AB038463], carboxypeptidase A2 (pancreatic) [BC007009], 2,4-dienoyl CoA reductase 2, peroxisomal [AE006463], protein kinase (cAMP-dependent, catalytic) inhibitor beta [AF225513], calpain 13 [AK027176], calpain 9 (nCL-4) [AF022799], fructose-1,6-bisphosphatase 1 [L10320], fructose-1,6-bisphosphatase [Y10812], gastric lipase [X05997], heat shock 10 ku protein [AJ250915], progastricsin (pepsinogen C) [M23077], protein tyrosine phosphatase, receptor type, N polypeptide 2 [U66702], carboxypeptidase A2 (pancreatic) [U19977], prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) [M59979], hyaluronoglucosaminidase 1 [U03056], galactose-4-epimerase, UDP-[L41668], chaperonin containing TCP1, subunit 3 (gamma)[BC008019], proprotein convertase subtilisin/kexin type 7 [BC010696], ubiquitin specific protease 14 (tRNA-guanine transglycosylase), [BC003556], Kallikrein 11 [AB013730], KIAA0089 protein [BC006168], prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) [S36219], prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) [U63846] |
| Metabolism |
| Aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) [AB032150], alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide [X76342], carbonic anhydrase II [J03037], carbonic anhydrase IX [Z54349], transcription termination factor, mitochondrial [Y09615], hydroxyprostaglandin dehydrogenase 15-(NAD) [L76465], dehydrogenase/reductase (SDR family) member 9 [AY017349], sulfotransferase family, cytosolic, 1C, member 1 [AF186261], ribonuclease, RNase A family, 4 [BC015520] |
| Organogenesis |
| GATA binding protein 4 [L34357], aldehyde dehydrogenase 3 family, member A2 [U46689], tropomyosin 4 [BC002827] |
| Regulation of blood pressure |
| Chromogranin A (parathyroid secretory protein 1) [BC009384] |
| Regulation of coagulation |
| Annexin A10 [AJ238979] |
| Regulation of metabolism |
| Estrogen-related receptor gamma [AB020639], zinc finger protein 345 [X78933], Kruppel-like factor 2 (lung) [AF134053], RAR-related orphan receptor C [U16997], PEPP subfamily gene 2 [AL590526] |
| Response to stress |
| Checkpoint suppressor 1 [BC007506], CHK1 (checkpoint, S.pombe) homolog [AF016582], for protein disulfide isomerase-related [D49490], glutathione S-transferase A3 [BG573805], glutathione synthetase [AK000947], glutathione S-transferase A4-4 (GSTA4) [AF025887], cathepsin E (CatE gene), [AJ250716], glutathione peroxidase 3 (plasma) [D00632], RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) [D14134], RAD51 (S. cerevisiae) homolog C [BC000667] |
| Signal transduction |
| G protein-coupled receptor 30[U63917], hypothetical protein FLJ22595[AK026248], G protein-coupled receptor 30[AF015257], RAP1, GTPase activating protein 1[M64788], active BCR-related gene[U01147], G protein-coupled receptor, family C, group 5, member B[AL137684], G protein-coupled receptor, family C, group 5, member C[AF207989], RAS protein activator like 1 (GAP1 like)[AF086713], regulator of G-protein signaling 17 [RGS17] mRNA, complete cds. [AF202257], histidine triad nucleotide-binding protein, [BC007090], RAB26, member RAS oncogene family, [BC007681] |
| Unclassified |
| Actin binding LIM protein 1 [NM006719], activated leucocyte cell adhesion molecule [AI050952], adrenergic, beta-2-, receptor, surface [NM000024], alcohol dehydrogenase 1C (class I), gamma polypeptide[NM000669], aldehyde dehydrogenase 1 family, member A1 [AV649527], aldehyde dehydrogenase 3 family, member A2 (BF679509), aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) [BI759009], annexin A7 [NM004034], arylacetamide deacetylase (esterase) [L32179], ATPase, H+ transporting, lysosomal (vacuolar proton pump) 42 ku [N75606], ATPase, H+/K+ exchanging, beta polypeptide [BI760752], ATP-binding cassette, sub-family C (CFTR/MRP), member 5 [NM005688], bA438B23.1 (neuronal leucine-rich repeat protein) [CAC22713], bB379O24.1 (novel protein similar to transcription factor GATA-5) [CAC36001], beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) [BE550952], calpain 2, (m/II) large subunit [BI771158], cathepsin E [AI598121], CDC28 protein kinase 2 [BG618998], chaperonin containing TCP1, subunit 2 (beta) [N98764], chromosome 1 open reading frame 8[BG476131], clathrin, light polypeptide (Lcb) (BE791502), cleavage and polyadenylation specific factor 5 25 ku subunit [AA738354], contactin 2 (axonal) [AI366526], cytochrome P450 isoform 4F12 [NM023944], cytochrome P450, subfamily IIC (mephenytoin 4-hydroxylase), polypeptide 9[BG567504], keratin 20 [X73501], delta sleep inducing peptide, immunoreactor [AL525317], DNA segment, numerous copies, expressed probes (GS1 gene) [AL570791], ESTs [AW960145], ESTs[N24233], ESTs, Highly similar to A32915 nucleophosmin [BG033160], ESTs, highly similar to AF078844 1 hqp0376 protein [BG165945], ESTs, Weakly similar to CA18 MOUSE COLLAGEN ALPHA 1(VIII) CHAIN PRECURSOR [M. musculus] [AW275800], ESTs, weakly similar to JC5314 CDC28/cdc2-like kinase associating arginine-serine cyclophilin [H. sapiens] [R85437], Fc fragment of IgG binding protein [D84239], flap structure-specific endonuclease 1 [BG773959], forkhead box O3A [AU134033], Friedreich ataxia region gene X123 [BI771919], galactose-4-epimerase, UDP-[BE388744], GATA-binding protein 6 [NM005257], glucosaminyl (N-acetyl) transferase 2, I-branching enzyme [NM001491], glutamic-pyruvate transaminase (alanine aminotransferase) [NM005309], glutathione S-transferase A4 [BI597618], granuphilin-a [BAA84656], hepatocyte nuclear factor 3, gamma [NM004497], highly expressed in cancer, rich in leucine heptad repeats [AA878068], holocytochrome c synthase (cytochrome c heme-lyase) [AL561481], Homer, neuronal immediate early gene, 2(BG742654), Homo sapiens clone 23763 unknown mRNA, partial cds [BG743005], homo sapiens SNC73 protein [SNC73] mRNA, complete cds[BF663123], homolog of yeast long chain polyunsaturated fatty acid elongation enzyme 2 [BF966630], human (clone HUAB2-3) Ig mRNA, variable region, partial cds. [L19893], human [JER47] MUC5AC mRNA for mucin (partial). [Z34277], peroxisomal membrane protein 2 22 ku (AIBC1) [AF250136], breast carcinoma amplified sequence 1 (AIBC1) [AF041260], Human cDNA FLJ11341 fis, clone PLACE1010786. [AK002203], human cDNA: FLJ20990 fis, clone CAE01666.[AK024463], cell-type T-cell Ig gamma-chain, V region (IGHV@) [L03144], KFAL6 lambda 1 Ig light chain variable region [AF124182], hypothetical protein FLJ22795 [AF316855], defensin, theta 1 [U10267], immunoglobulin heavy diversity 2-21 [AB019441], Human gastric H,K-ATPase catalytic subunit gene, complete cds. [M63962], genomic DNA of 21q22.2 Down Syndrome region, segment 11/13. [AP000020], BIC transcript [AP001693], immunoglobulin kappa variable 1-8 [AP001209], genomic DNA, chromosome 8q23, clone:KB1747F8. [AP003113], glucocorticoid receptor alpha mRNA, variant 3’ UTR [U25029], immunoglobulin lambda constant 1 (Mcg marker) [D87023], Ig lambda light chain variable region gene (24-12ITIIIE213) rearranged; Ig-Light-Lambda; VLambda. [Z85038], Ig rearranged gamma chain mRNA, V-J-C region and complete cds. [M63438], Ig rearranged gamma chain mRNA, V-J-C region and complete cds. [M63438], metallothionein 1F (functional) [M13003], metallothionein 1A (functional) [K01383], mRNA for anti-Sm antibody VL chain (V kappa 4/J kappa 3). [Z46347], heat shock 70 ku protein 4 [AB023420], Ig heavy chain variable region, clone ToPA214. [Z98733], Ig kappa light chain, anti-RhD, therad 7. [AJ010422], Ig lambda light chain. [Y14738], sterile alpha and TIR motif containing 1 [AJ290445], KIAA0832 protein, complete cds.[AB020639], synaptotagmin-like 2 [AB046817], metallothionein 1L [X97261], NPC-related protein NAG73 [AF280797], prion protein (p27-30) (Creutzfeld-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) [AY008282], Human promyelocytic leukemia zinc finger protein (PLZF) gene, complete cds. [AF060568], cystatin A (stefin A) [X05978], sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F [U38276], serine protease inhibitor, Kazal type 1 [AF286028], SM22 alpha gene, 5’ flanking region. [D84344], hypothetical protein MGC27165 [AF067420], activating transcription factor 4 pseudogene (tax-responsive enhancer element B67 pseudogene) [U03712], chromosome 7 open reading frame 28B [BC010130], Human, clone MGC:2392 IMAGE:2961444, [BC001646], Human, LIM domain only 4, clone MGC:872 IMAGE:3355972, [BC003600], S100 calcium binding protein P [BC006819], Similar to Ig kappa constant, clone MGC:12418 IMAGE:3934658, [BC005332], hyaluronoglucosaminidase 1 [AL578077], hypothetical protein [AI202106], immunoglobulin heavy constant mu [BM008087], immunoglobulin heavy constant mu [AI634950], immunoglobulin lambda joining 3 [BF338816], immunoglobulin lambda locus [M87790], IMP (inosine monophosphate) dehydrogenase 1[AL518727], interleukin 1, beta [AA577318], karyopherin alpha 2 (RAG cohort 1, importin alpha 1) [BE889289], KIAA0008 gene product [BI087140], KARP-1-binding protein [AB022659], KIAA0657 protein [BC007201], family with sequence similarity 13, member A1 [AF009202], glutamate receptor interacting protein 2 [AF052177], KIAA1727 protein [AB051514], Kruppel-like factor 4 (gut)[AI568487], lactotransferrin [BI021407], lamin B receptor [L25931], lethal giant larvae homolog 2 (Drosophila) [AK025401], leukemia inhibitory factor receptor [NM002310], lipase, gastric [NM004190], Macaque dd-4 gene for 3(20)alpha-hydroxysteroid/dihydrodiol dehydrogenase, complete cds. [AB020711], Macaque testis cDNA clone:QtsA-14970, full insert sequence. [AB070147], mammaglobin 2 [NM002407], membrane-bound transcription factor protease, site 2 [AU099021], metallothionein 1E (functional) [H72532], metallothionein 1X[BF130769], methylmalonate-semialdehyde dehydrogenase [BG743099], minichromosome maintenance deficient (S. cerevisiae) 2 (mitotin) [BG491883], mucin 5, subtypes A and C, tracheobronchial/gastric [AW867962], myeloid differentiation primary response gene (88) [AW965179], myosin X [AU151619], non-metastatic cells 1, protein [NM23A] expressed in [BG753664], osteoblast specific factor 2 (fasciclin I-like) [N71912], pancreatitis-associated protein [NM002580], peptidylprolyl isomerase F (cyclophilin F) [AL531948], period (Drosophila) homolog 1 [BE615751], phosphatidic acid phosphatase type 2B [AW131816], phosphoinositide-3-kinase, class 2, gamma polypeptide [BG196286], phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) [BE884737], pituitary tumor-transforming 1 [AW957275], platelet-derived growth factor receptor, alpha polypeptide [AW887370], plexin B2 [BC004542], potassium inwardly-rectifying channel, subfamily J, member 15 [BG288548], predicted using Genefinder~contains similarity to Pfam domain: PF00465 (Iron-containing alcohol dehydrogenases), Score=177.7, E-value=1.9e-50, N=2~cDNA EST EMBL:Z14517 comes from this gene; cDNA EST yk18d4.3 comes from this gene~c [CAA21631], proprotein convertase subtilisin/kexin type 7 [BG820292], prostate stem cell antigen (AJ297436), protein disulfide isomerase [NM006849], protein tyrosine phosphatase, receptor-type, Z polypeptide 1 [BI488535], patative [BAB32258], RAB27A, member SAS oncogene family [AL120794] RAD21 (S. pombe) homolog [BF696386], RAP1, GTPase activating protein 1 [M64788], Ras homolog enriched in brain 2 [AW519065], RecQ protein-like 5 [AI123482], regenerating islet-derived 1 alpha (pancreatic stone protein, pancreatic thread protein) [BI713022], replication factor C (activator 1) 2 (40 ku) [BE295474], replication factor C (activator 1) 5 (36.5 ku) [AL525427], ribonuclease, RNase A family, 1 (pancreatic) [BI596306], ribosomal protein L12 [BG471683], ribosomal protein S10 [BC004334], ribosomal protein S12 [AA314429], ribosomal protein S15a [BG285655], ribosomal protein S20 [BG684583], S100 calcium-binding protein P [AI148603], pleckstrin homology domain containing, family E (with leucine rich repeats) member 1 [AB011178], selenium binding protein 1 [BC009084], serine/threonine kinase 15 [AI038260], Sjogren syndrome antigen B (autoantigen La) [AA479280], small nuclear ribonucleoprotein D2 polypeptide (16.5 ku) [BM015197], sodium channel, nonvoltage-gated 1 alpha [BI160595], sodium channel, nonvoltage-gated 1, beta (Liddle syndrome) [BI763841], sodium channel, nonvoltage-gated 1, beta (Liddle syndrome) [AI683977], somatostatin [BI713744], stem-loop (histone) binding protein [AW150631], trefoil factor 2 (spasmolytic protein 1) [BI517365], tumor necrosis factor (ligand) superfamily, member 13BG [272792], UDP-glucose ceramide glucosyltransferase [AI609116], vaccinia related kinase 1 [AA312869], v-akt murine thymoma viral oncogene homolog 2 [AU130605], vascular cell adhesion molecule 1 [AL037837] |
The completion of human genome sequencing has facilitated high-throughput quantitative analysis of gene expression alterations. Such transcriptome analyses utilizing DNA microarray offer a new avenue to understand the biological diversity of human cells and tumors. Microarray analysis of malignancies from other organ sites has revealed molecular subtypes of tumors that are histologically indistinguishable and clinically informative[26-29]. To elucidate the molecular portrait of GC, we extracted RNA from microdissected normal gastric epithelium and corresponding tumor cells for examining gene expression profiles in the cells as they exist in vivo. Coupling with common “reference” sample as an internal standard, we found this strategy is a rapid and efficient way to identify differentially expressed genes and can readily distinguish GC into 3 molecular subtypes.
Most previous gene expression studies on GCs have used whole tumor tissues[10-21]. The overall information of genetic alterations with this approach was comprehensive but might be hampered by the fact that stromal and inflammatory cells usually intermingled with cancer cells. Therefore, special procedures to isolate cancer cells from heterogeneous tissues are mandatory. To overcome the obstacles of the contamination of cancer cells with wrong cells, we adopted LCM. LCM allows for the rapid, reliable and accurate procurement of cells from the specific microscopic regions of tissue sections under direct visualization. The proportion of contaminated cells with this method is estimated to the less than 0.3%[30]. It thus affords the opportunity to perform molecular genetic analysis of pure populations of malignant cells in their native tissue environment.
However, the RNAs obtained from procured cancer cells were not sufficient to hybridize with cDNA microarray. The recent advances of amplification techniques have provided the key to resolve this problem[31]. In particular, linear RNA amplification could not only provide sufficient RNAs for further analyses but also preserve the overall genetic information.
By the successfully developed techniques of LCM, linear amplification of RNA and cDNA microarray, we found that 504 genes could distinguish cancerous and non-cancerous groups with more than 99% accuracy. Of the genes discovered, some have known association with gastrointestinal cancers. Among them, trefoil factors 1, 2, and 3 were in the list. Furthermore, the expression patterns of these 3 genes were in agreement with previous reports in GCs[32]. These results confirmed our microarray data indirectly. Moreover, we selected trefoil factor 1 for further verification by immunohistochemical staining and the data were also consistent. The remaining novel genes differentially expressed in cancerous and non-cancerous tissues provided a starting point for identification of new biomarkers in GC and are worthy of future in-depth studies to elucidate their roles in GCs.
By comparing expression patterns of the identified 504 genes in individual tissues, we noticed a phylogenetic tree that not only showed clear segregation between normal and cancerous gastric tissues but also assorted GC into 3 subtypes. Although most previous studies suggested classification of GC based on gene expression largely recapitulate that based on histology[33], we did not find such correlation in our system. This discrepancy may be, in part, due to methodological differences and small case numbers in our study. In addition, one intriguing possibility is that tumors sharing similar genes expression profile arise from a common molecular genetic lesion but present with different histological appearances[16,18]. Enrollment of a larger number of tumor samples with well-documented information is under way to further clarify the clinicopathological significance of this molecular classification.
Another unique feature distinct from previous microarray studies of GC is that we hybridized each tumor or normal mRNA against a “universal reference” of mRNA. This concept was advocated by investigators from Stanford University, who prepared cDNA for microarray experiments from a pool of mRNA isolated from 11 different cells lines[29]. Although normal epithelium from a spectrum of histological changes from atrophy to metaplasia could be easily microdissected by LCM, the so-called normal epithelium adjacent to tumor may not be similar to epithelium distant from the tumor at the molecular level. Therefore, the definition of control normal tissue will need to be carefully assessed in future global analysis of gene expression.
In conclusion, combined LCM and cDNA microarray is a rapid and efficient way to identify differentially expressed genes and can readily distinguish GCs by their molecular portraits. Further characterization of the genes identified in this study and prospective translational studies to determine their potential clinical value may lead to a deeper insight into the pathogenesis of GCs and facilitate the development of novel biomarkers for diagnostic and therapeutic applications.
Science Editor Kumar M and Guo SY Language Editor Elsevier HK
| 1. | Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 Suppl 8:S4-S66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1370] [Cited by in RCA: 1370] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 2. | Wu MS, Lin JT, Lee WJ, Yu SC, Wang TH. [Gastric cancer in Taiwan]. J Formos Med Assoc. 1994;93 Suppl 2:S77-S89. [PubMed] |
| 3. | Lin JT, Wu MS, Wang JT, Shun CT, Chen CJ, Wang TH. Clinicopathologic study of 208 patients with early gastric cancer in Taiwan: a comparison between Eastern and Western countries. J Gastroenterol Hepatol. 1994;9:344-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Fuchs CS, Mayer RJ. Gastric carcinoma. N Engl J Med. 1995;333:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 442] [Article Influence: 14.7] [Reference Citation Analysis (1)] |
| 5. | Wright PA, Quirke P, Attanoos R, Williams GT. Molecular pathology of gastric carcinoma: progress and prospects. Hum Pathol. 1992;23:848-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Stemmermann G, Heffelfinger SC, Noffsinger A, Hui YZ, Miller MA, Fenoglio-Preiser CM. The molecular biology of esophageal and gastric cancer and their precursors: oncogenes, tumor suppressor genes, and growth factors. Hum Pathol. 1994;25:968-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 80] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Tahara E, Semba S, Tahara H. Molecular biological observations in gastric cancer. Semin Oncol. 1996;23:307-315. [PubMed] |
| 8. | Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1818] [Cited by in RCA: 1693] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 9. | Alizadeh AA, Ross DT, Perou CM, van de Rijn M. Towards a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 185] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 10. | Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233-240. [PubMed] |
| 11. | Ji J, Chen X, Leung SY, Chi JT, Chu KM, Yuen ST, Li R, Chan AS, Li J, Dunphy N. Comprehensive analysis of the gene expression profiles in human gastric cancer cell lines. Oncogene. 2002;21:6549-6556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Sakakura C, Hagiwara A, Nakanishi M, Shimomura K, Takagi T, Yasuoka R, Fujita Y, Abe T, Ichikawa Y, Takahashi S. Differential gene expression profiles of gastric cancer cells established from primary tumour and malignant ascites. Br J Cancer. 2002;87:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Inoue H, Matsuyama A, Mimori K, Ueo H, Mori M. Prognostic score of gastric cancer determined by cDNA microarray. Clin Cancer Res. 2002;8:3475-3479. [PubMed] |
| 14. | Weiss MM, Kuipers EJ, Postma C, Snijders AM, Siccama I, Pinkel D, Westerga J, Meuwissen SG, Albertson DG, Meijer GA. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene. 2003;22:1872-1879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Boussioutas A, Li H, Liu J, Waring P, Lade S, Holloway AJ, Taupin D, Gorringe K, Haviv I, Desmond PV. Distinctive patterns of gene expression in premalignant gastric mucosa and gastric cancer. Cancer Res. 2003;63:2569-2577. [PubMed] |
| 16. | Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309-3316. [PubMed] |
| 17. | Suganuma K, Kubota T, Saikawa Y, Abe S, Otani Y, Furukawa T, Kumai K, Hasegawa H, Watanabe M, Kitajima M. Possible chemoresistance-related genes for gastric cancer detected by cDNA microarray. Cancer Sci. 2003;94:355-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Kim B, Bang S, Lee S, Kim S, Jung Y, Lee C, Choi K, Lee SG, Lee K, Lee Y. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248-8255. [PubMed] |
| 19. | Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004;10:272-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 20. | Kim HK, Choi IJ, Kim HS, Kim JH, Kim E, Park IS, Chun JH, Kim IH, Kim IJ, Kang HC. DNA microarray analysis of the correlation between gene expression patterns and acquired resistance to 5-FU/cisplatin in gastric cancer. Biochem Biophys Res Commun. 2004;316:781-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Nørsett KG, Laegreid A, Midelfart H, Yadetie F, Erlandsen SE, Falkmer S, Grønbech JE, Waldum HL, Komorowski J, Sandvik AK. Gene expression based classification of gastric carcinoma. Cancer Lett. 2004;210:227-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome-wide analysis of gene expression in intestinal-type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res. 2002;62:7012-7017. [PubMed] |
| 23. | Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39-S47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Haraguchi N, Inoue H, Mimori K, Tanaka F, Utsunomiya T, Yoshikawa K, Mori M. Analysis of gastric cancer with cDNA microarray. Cancer Chemother Pharmacol. 2004;54 Suppl 1:S21-S24. [PubMed] |
| 25. | Wu MS, Shun CT, Wang HP, Lee WJ, Wang TH, Lin JT. Loss of pS2 protein expression is an early event of intestinal-type gastric cancer. Jpn J Cancer Res. 1998;89:278-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 997] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 27. | Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7015] [Cited by in RCA: 6305] [Article Influence: 252.2] [Reference Citation Analysis (0)] |
| 28. | Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1199] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 29. | Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406:747-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10665] [Cited by in RCA: 10943] [Article Influence: 437.7] [Reference Citation Analysis (0)] |
| 30. | Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385-2400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 197] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 31. | Aoyagi K, Tatsuta T, Nishigaki M, Akimoto S, Tanabe C, Omoto Y, Hayashi Si, Sakamoto H, Sakamoto M, Yoshida T. A faithful method for PCR-mediated global mRNA amplification and its integration into microarray analysis on laser-captured cells. Biochem Biophys Res Commun. 2003;300:915-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Katoh M. Trefoil factors and human gastric cancer (review). Int J Mol Med. 2003;12:3-9. [PubMed] |
| 33. | Jinawath N, Furukawa Y, Hasegawa S, Li M, Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H. Comparison of gene-expression profiles between diffuse- and intestinal-type gastric cancers using a genome-wide cDNA microarray. Oncogene. 2004;23:6830-6844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |