Published online Sep 14, 2005. doi: 10.3748/wjg.v11.i34.5277
Revised: November 14, 2004
Accepted: November 19, 2004
Published online: September 14, 2005
AIM: To study the effects of Pinus massoniana bark extract (PMBE) on cell proliferation and apoptosis of human hepatoma BEL-7402 cells and to elucidate its molecular mechanism.
METHODS: BEL-7402 cells were incubated with various concentrations (20-200 µg/mL) of PMBE for different periods of time. After 48 h, cell proliferation was determined by 3-(4,5-dimethyl-thiazolyl-2)-2,5-diphenyl tetrazolium bromide (MTT) assay. Apoptosis was evaluated by morphological observation, agarose gel electrophoresis, and flow cytometry analysis. Possible molecular mechanisms were primarily explored through immunohistochemical staining.
RESULTS: PMBE (20-200 µg/mL) significantly suppressed BEL-7402 cell proliferation in a time- and dose-dependent manner. After treatment of BEL-7402 cells with 160 µg/mL PMBE for 24, 48, or 72 h, a typical apoptotic “DNA ladder” was observed using agarose gel electrophoresis. Nuclear condensation and boundary aggregation or split, apoptotic bodies were seen by fluorescence and electron microscopy. Sub-G1 curves were displayed by flow cytometry analysis. PMBE decreased the expression levels of Bcl-2 protein in a time-dependent manner after treatment of cells with 160 µg/mL PMBE.
CONCLUSION: PMBE suppresses proliferation of BEL-7402 cells in a time- and dose-dependent manner and induces cell apoptosis by possibly downregulating the expression of the bcl-2 gene.
- Citation: Cui YY, Xie H, Qi KB, He YM, Wang JF. Effects of Pinus massoniana bark extract on cell proliferation and apoptosis of human hepatoma BEL-7402 cells. World J Gastroenterol 2005; 11(34): 5277-5282
- URL: https://www.wjgnet.com/1007-9327/full/v11/i34/5277.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i34.5277
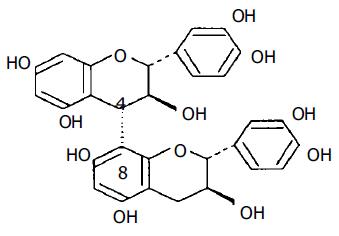
Pinus massoniana needles and bark have been commonly used in diet therapies in eastern Asia (mainly China and Japan) for thousands of years, but until the last decade of the 20th century, bioactive substances from P massoniana bark have not been successfully extracted through crude sieving, centrifugation, or micro- and ultra-filtration with hollow fiber and coiled-spiral nanofiltration membranes[1]. Standard PMBE contains 26.0-28.3% flavonoids (mainly procyanidins) as important bioactive substances. The most important of these are included in the procyanidin B series, which only differ in the number and position of hydroxyl groups, as can be seen for procyanidin B3 in Figure 1[2].
PMBE is well known for its antioxidant properties, which may result from its ability to scavenge free radicals[1]. It is this antioxidant activity that may be helpful in the prevention and therapy of diseases and degenerative processes associated with oxidative stress. The bark of P massoniana has been mentioned in the traditional Chinese pharmacopoeia, and has been mainly used to promote hemostasis and detoxification, and for the treatment of rheumatic arthralgia, hypertension, and chilblain in China and other countries of the Orient[3]. Our previous research results showed that PMBE can broadly inhibit the growth of eight different cancer cell lines in vitro[4]. Additionally, we found that PMBE can selectively inhibit the proliferation of human hepatoma BEL-7402 cells without impacting the growth of normal human liver L-02 cells at an effective concentration range similar to that of the chemotherapeutic agent, cisplatin[4]. It may play a role in cancer clinic as both antidote and potentiator of surgical therapy.
Considering that hepatoma is the most common malignant tumor occurring in both men and women in China, we wished to assess the processes by which PMBE inhibits growth and affects apoptosis of BEL-7402 cells and elucidate a possible molecular mechanism for these actions. It is reported that extracts from P maritima bark (pycnogenol)[5] and from P radiata bark (enzogenol)[6] have been used therapeutically in southwestern Europe (mainly France, Germany, and Spain), Australia, and New Zealand for nearly 10 years. Huynh and Teel[7] once reported that pycnogenol selectively induces apoptosis of human mammary cancer cells (MCF-7), but does not induce apoptosis of normal human mammary MCF-10 cells. It is reasonable to hypothesize that PMBE may also be helpful in therapies of human hepatoma, possibly through the regulation of cell proliferation and/or cell death. Therefore, this study aimed to assess the processes by which PMBE inhibits cancer cell growth, affects apoptosis of BEL-7402 cells, and the possible molecular mechanism for these actions.
PMBE was provided by the Institute of Songzhen (Guangdong Province, China). A stock solution of 10 mg/mL PMBE was made in 100% dimethylsulfoxide (DMSO) and filtered via a 0.22 µm minipore membrane. The stock solution was aliquoted and stored at -20°C, thawed just before testing, and diluted with the culture medium. The final concentration of DMSO for all treatments was less than 0.25%.
Human hepatoma BEL-7402 cells were purchased from the Animal Cell Center, Medical College, Sun Yat-Sen University, China. BEL-7402 cells were incubated in RPMI 1640 medium (Sijiqing Co., Zhejiang, China) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, 100 µg/mL streptomycin, and 10 mmol/L HEPES, pH 7.4, and kept at 37°C in a humidified 50 mL/L CO2 incubator. For this study, cells were either incubated in the absence or presence of different concentrations of PMBE for 48 h or exposed to 160 µg/mL PMBE for different time periods.
Growth inhibition of BEL-7402 cells by PMBE was measured by 3-(4,5-dimethyl-thiazolyl-2)-2,5-diphenyl tetrazolium bromide (MTT) assay[8,9] with minor modifications. Briefly, cancer cells were seeded into 96-well microtiter plates at appropriate densities to maintain the cells in an exponential phase of growth during the experiment. BEL-7402 cells were exposed to PMBE either at 20, 40, 80, 160, or 200 µg/mL for 48 h or to 160 µg/mL PMBE for either 24, 48, or 72 h. Each concentration was tested in quadruplicate. At the end of the treatment, 20 µL of 5 mg/mL MTT (Sigma, St. Louis, MO, USA) was added to each well and the plates were incubated for 4 h at 37°C. DMSO (150 µL) was then added to each well and the plates were rotated for 10-20 min at 200 r/min by a shaker (THZ-C, Shenhua BioTech Co., Shenzhen, China). The optical absorbance was read on a plate reader (BIO-RAD Co., USA) at a wavelength of 570 nm. Media and DMSO controls were included for all experiments. The inhibitory rate of cell proliferation was calculated using the following formula: Growth inhibition (%) = (Acontrol-Atreated)/Acontrol × 100. Results were measured in triplicate.
Fluorescence microscopy BEL-7402 cells were seeded in sterile culture dishes on coverslips at a density of 105 cells/mL and cultured in 10% FBS-RPMI 1640 medium at 37°C in a humidified atmosphere of 50 mL/L CO2. Cells were grown to 80-90% confluence and synchronized by incubation in 0.5% FBS-RPMI 1640 for 56 h. The experiments were divided into test and control groups. Cells in the test group were incubated in 10% FBS-RPMI 1640 medium containing 160 µg/mL PMBE for 24 or 48 h. Cells in the control group were incubated in 10% FBS-RPMI 1640 alone for 24 h. The coverslips were washed with PBS (containing Ca2+, Mg2+) for 3-5 min, then a 1 000:1 mixture of methanol: DAPI was added and placed at 4°C for 15-30 min, and then washed thrice with PBS for 3-5 min to reduce the background signal. Finally, the cells fixed on the coverslips were analyzed, and photographed under fluorescence microscope (BH-2, Olympus, Japan).
Electron microscopy BEL-7402 cells were incubated in 10% FBS-RPMI 1640 medium with or without 160 µg/mL PMBE at 37°C in a humidified atmosphere of 50 mL/L CO2 for 72 h. After treatment with 0.25% trypsin, 5 × 106 cells were collected, centrifuged at 10 000 g for 5 min and then washed twice with PBS for 5 min. Cell pellets were fixed in 2.5% glutaraldehyde in PBS at 4°C overnight. After washing thrice (10 min each) in PBS, the pellets were incubated in 1% osmium tetroxide (Electron Microscopy Sciences) in PBS for 1 h at room temperature. After three more 10-min washes in PBS, the pellets were dehydrated using a successive ethanol series of 300, 500, 700, 800, and 950 mL/L in double distilled water (10 min each) at room temperature. The pellets were further dehydrated at room temperature in 100% acetone (3 × 10 min), incubated in 50% acetone/50% epoxy resin (Durcupan ACM, EMS) at room temperature for 1 h, and infiltrated with 100% epoxy resin (3 × 1 h) at room temperature. After the final incubation, the resin-infiltrated pellets plus excess resin were poured into capsules and polymerized at 70°C overnight. Ultrathin (500 Å) sections were made using a heat-swelling ultramicrotome (LKB-NoVa-V, Switzerland). After being stained with uranium acetate and plumbum citrate, the sections were observed under a transmission electron microscope (JEM 100CX-II, Houghton, MI, USA) and electron micrographs were taken.
BEL-7402 cells were seeded in two 25-mL flasks at a density of 105 cells/mL and incubated in 10% FBS-RPMI 1640 medium as previously described in the presence or absence of 160 µg/mL PMBE for 24 h. After harvesting the cells, samples were washed with PBS and centrifuged at 1 000 r/min. The cell pellets were resuspended in a 40-µL solution of 0.2 mol/L NaH2PO4/0.1 mol/L citric acid (192:8) and incubated at room temperature for 60 min. After centrifugation for 30 min at 480 r/min, the supernatant was transferred to a 1.5 mL microcentrifuge tube. Three microliters of NP-40 (2.5 mg/mL) and 3 µL RNase A (10 mg/mL) were added to the tube and incubated at 37°C for 60 min. After the addition of 3 µL proteinase K (10 mg/mL), the tube was once again incubated at 50°C for 50 min. At the end of the reaction, 5 µL loading buffer was added and samples were electrophoresed on a 10 g/L agarose gel containing 0.5 µg/mL ethidium bromide, at 7.5 V/cm in 1 × TAE buffer for 1.5 h. Analyses and photography were performed with the gel imaging and analysis system (BIO-RAD Co., USA).
Cellular DNA content and cell distribution were quantified by flow cytometry using propidium iodide (PI)[11]. After treatment with PMBE, the cells were collected, washed in PBS, and fixed in 700 mL/L ethanol at 4°C overnight. After fixation, the cells were washed in PBS and pellets obtained by centrifugation were stained with PI staining solution containing 50 µg/mL PI, 10 µg/mL RNase A, 5 mL/L Triton X-100, and 0.1% (w/v) trisodium citrate for 30 min at room temperature in the dark. DNA content was determined by flow cytometry (ELITE, BECKMAN Co., USA). Cell populations in the sub-G1 area (the position where apoptotic cells located) were quantified from a standard count of 104 cells using MULTYCYCLE software (PHEONIX Co., USA).
BEL-7402 cells were seeded in sterile culture dishes on coverslips at a density of 105 cells/mL, and incubated as described previously. The experiments were divided into test and control groups. Cells in the test group were incubated with 10% FBS-RPMI 1640 medium containing 160 µg/mL PMBE for 24, 48, or 72 h. Cells in the control group were incubated with 10% FBS-RPMI 1640 alone for 24 h. Cells were first washed thrice (1 min each) with PBS, fixed in acetone for 10 min, immersed in 0.75% H2O2-PBS at 37°C for 30 min to inactivate the peroxidase, and washed twice with PBS for 3 min. Normal goat serum was used to eliminate the unspecific staining. Fixed cells were incubated for 30 min at room temperature with mouse anti-human Bcl-2 mAb (primary antibody, Oncogene Research Products) and washed five times with PBS. Slides were then incubated at 37°C for 30 min with biotinylated goat anti-mouse IgG (secondary antibody), washed twice with PBS, and visualized using the immunoperoxidase-based SABC kit (Boster Co.) under a phase contrast microscope. According to the kit protocol, BEL-7402 cells treated with 160 µg/mL of PMBE for 24 h without the addition of primary antibody were used as the negative control. Cells were regarded as positive, when the cell membrane was stained in a brown coffee color similar to the positive control provided by the kit manufacturer.
Data were expressed as mean±SD. All experiments were performed in triplicate, unless otherwise indicated. Statistical significance was evaluated either by Student’s t-test or by one-way ANOVA using SPSS10.0 software (P < 0.05 was considered statistically significant, P < 0.01 very statistically significant).
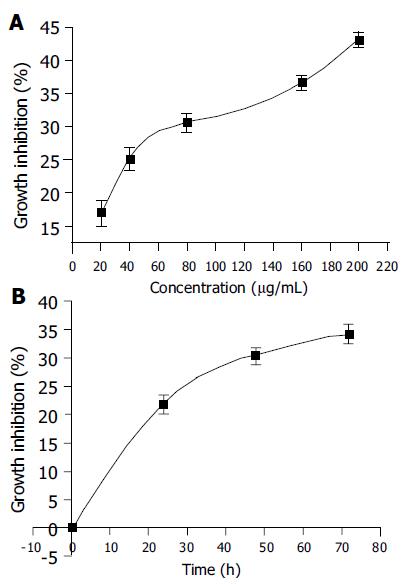
Human hepatoma BEL-7402 cells were incubated with PMBE for different periods of time at concentrations ranging 20-200 µg/mL and analyzed by MTT assay. PMBE was shown to inhibit the proliferation of BEL-7402 cells in a dose- and time-dependent manner. After exposure of BEL-7402 cells to 20, 40, 80, 160, or 200 µg/mL of PMBE for 48 h, cell proliferation was inhibited by 16.97%, 25.20%, 30.64%, 36.64%, and 43.17%, respectively (Figure 2A). Treatment of BEL-7402 cells with 160 µg/mL PMBE for 24, 48, or 72 h, inhibited cell proliferation by 21.96%, 30.47%, and 34.36%, respectively (Figure 2B).
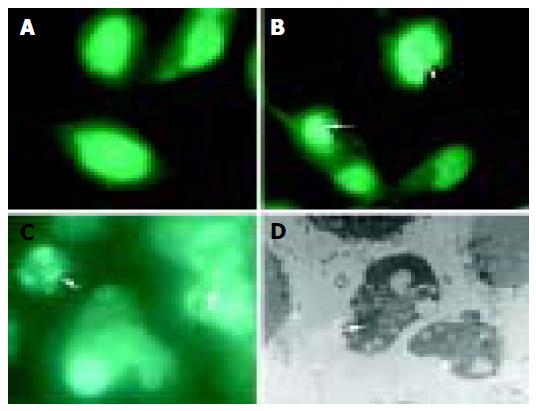
Under fluorescence microscope, untreated BEL-7402 cells displayed extended, flat cell bodies with uniform chromatin in the nuclei (Figure 3A). The nuclei of BEL-7402 cells, treated with 160 µg/mL PMBE for 24 h, showed the characteristic morphological changes of apoptosis, including nuclear condensation, boundary aggregation and split, even DNA fragmentation (Figure 3B), which dramatically increased over time (Figure 3C). Under transmission electron microscope, apoptotic bodies were observed in cells treated with 160 µg/mL PMBE for 72 h (Figure 3D).
In order to demonstrate that PMBE was able to induce an apoptotic response, we analyzed DNA degradation in PMBE-treated BEL-7402 cells by agarose gel electrophoresis. PMBE, at a concentration of 160 µg/mL, induced the typical inter-nucleosomal DNA fragmentation in multiples of 180-200 bp after 24 h, a hallmark of apoptosis (Figure 4). No chromosomal DNA cleavage was observed in the untreated control cells (Figure 4).
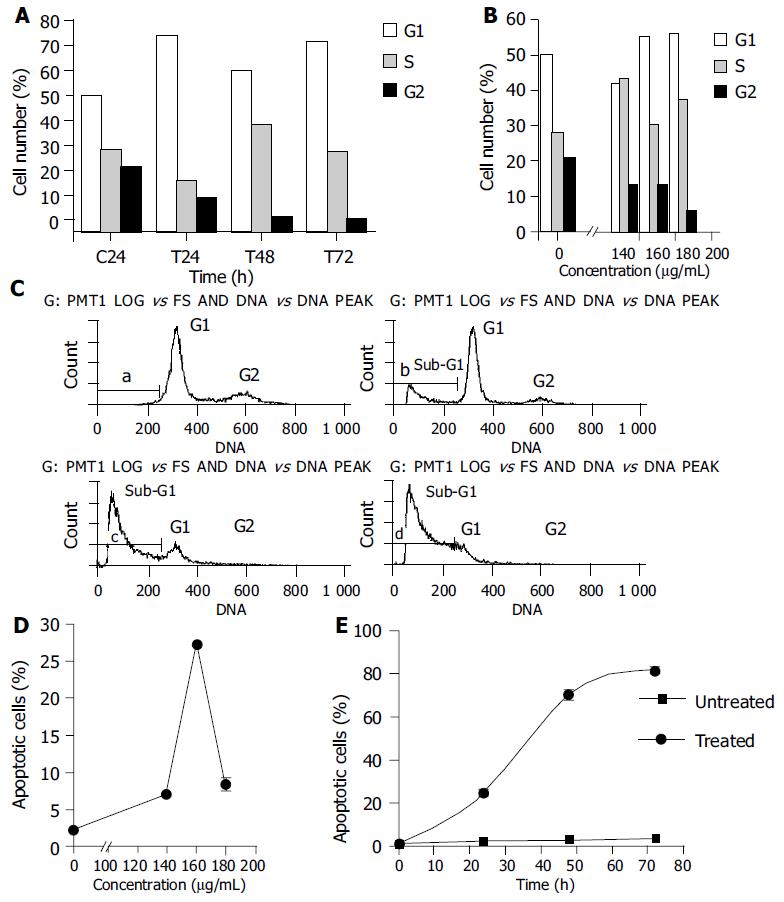
Apoptotic cells could be recognized by PI staining due to DNA degradation and subsequent leakage of stain from the cells. The hypodiploid population, formed by cells with a reduced DNA content in frequency histograms (sub-G1 peak), could be quantified by flow cytometry and effects of PMBE on BEL-7402 cell distribution could be observed. Results showed that incubation with 160 µg/mL of PMBE for different time periods or with 140, 160, or 180 µg/mL of PMBE for 24 h resulted in accumulation of cells at G1 or S phase (Figures 5A and B) and a blockade of cell proliferation. Arrest of the cell cycle at G1 or S phase eventually led to cell death, as the prolonged incubation with 160 µg/mL PMBE induced an increasing apoptotic response, resulting in the rapid appearance of cells with a DNA content less than that at G1 phase, characteristic of apoptotic cells (Figures 5C). In addition, PMBE induced apoptosis of BEL-7402 cells in a dose-dependent manner. In untreated BEL-7402 cells, the mean apoptotic population was 2.23%, which increased to 7.03%, 28.3%, and 8.43% after a 24-h treatment with 140, 160, and 180 µg/mL PMBE, respectively (Figure 5D). For the time-dependent relationship, the number of apoptotic cells increased with prolonged incubation in PMBE. The number of apoptotic cells increased from 2.23% to 25.1%, 70.6%, and 81.7% after 24, 48, and 72 h of exposure to 160 µg/mL PMBE, respectively (Figure 5E). Therefore, PMBE induced apoptosis of BEL-7402 cells in a dose- and time-dependent manner.
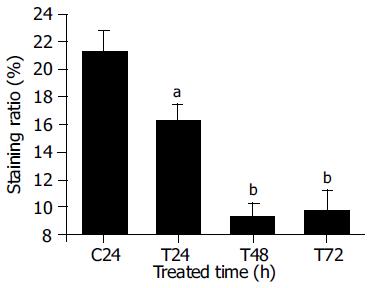
The involvement of the bcl-2 gene in apoptosis was reported in several studies of anticancer drugs[12-14]. The role of Bcl-2 in PMBE-induced apoptosis of human hepatoma BEL-7402 cells was evaluated by immunohistochemical analysis. Three hundred cells were observed under a microscope and the positive cell ratio was calculated from three independent experiments. The results showed that cells in the control group had higher expression levels of Bcl-2, with a (21.3 ± 1.5)% positive staining ratio, while Bcl-2 expression in test group cells decreased to (16.3 ± 1.2)%, (9.3 ± 0.9)%, and (9.7 ± 1.4)% after treatment with 160 µg/mL PMBE for 24, 48, or 72 h, respectively (Figure 6). Compared with control, all test groups displayed significant differences (P < 0.05). The positive staining ratio of Bcl-2 protein expression decreased with PMBE treatment time, suggesting that PMBE might play a role in inducing apoptosis of human hepatoma BEL-7402 cells by possibly down-regulating the expression of the bcl-2 gene.
Hepatoma is one of the most common malignant tumors in China and Asia. Therefore, we chose the human hepatoma cell line BEL-7402, as an in vitro model to explore the effects of PMBE on cancer cell growth and apoptosis to evaluate the value of PMBE in cancer prevention and therapy.
Our results showed that PMBE at doses ranging 20-200 µg/mL, significantly suppressed cell proliferation in a time- and dose-dependent manner and induced characteristic morphological and biochemical changes associated with apoptosis of human hepatoma BEL-7402 cells. The inhibitory effect of PMBE on cancer cell growth may be attributed to its antioxidant function[1]. Additionally, PMBE appears to induce apoptosis via downregulation of the Bcl-2 protein. Moreover, we have observed that PMBE exhibits more potent effects on apoptosis than on growth inhibition at similar concentrations as described in the results. When exposed to 160 µg/mL of PMBE for 24, 48, and 72 h, the growth inhibition ratio and apoptosis ratio of BEL-7402 cells were 21.96%, 30.47%, 34.36% and 25.1%, 70.6%, 81.7%, respectively. These distinct actions on cell cycle progression and apoptosis suggest that PMBE is able to trigger different signal transduction pathways, and to exert effects on two key processes during cancer treatment. Taken together, our data demonstrate PMBE has a high capacity of suppressing cell proliferation and triggering apoptosis of human hepatoma cells. Therefore, PMBE may have a therapeutic potential in human liver cancer and may find its use as an anticancer candidate.
The activities of PMBE may account for previously reported cytotoxic effects of this extract[4]. In this study, we showed compelling evidence for pro-apoptotic activity of PMBE using a combination of different techniques. Furthermore, these actions are time- and dose-dependent. We have also found that PMBE could induce accumulation of cells arrested at G1 and S phases of the cell cycle. Therefore, the antitumor effects of PMBE may result from multiple mechanisms of action, such as interfering with cell cycle progression and inducing apoptosis, possibly by decreasing Bcl-2 protein expression. Oncogenes such as bcl-2, are often involved in the development of tumors, and may be related to tumor cell drug resistance[15]. Our results showed that PMBE not only induced BEL-7402 cell apoptosis, but also drastically decreased Bcl-2 protein expression concurrently. The identification of the genes involved and their related signal transduction pathways critical to tumorigenesis await further study.
In conclusion, PMBE is capable of inhibiting growth and inducing apoptosis of human hepatoma BEL-7402 cells in a dose- and time-dependent manner, indicating that PMBE may hold promise as an anticancer agent.
We thank the reviewers for their helpful comments.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Cui YY, Xie H, Lai F, Wang JF. Preliminary study on antioxidative activities of Pinus massoniana bark extract (PMBE). Shipin Kexue. 2004;25:179-183. |
| 2. | Zhao J, Wang J, Chen Y, Agarwal R. Anti-tumor-promoting activity of a polyphenolic fraction isolated from grape seeds in the mouse skin two-stage initiation-promotion protocol and identification of procyanidin B5-3'-gallate as the most effective antioxidant constituent. Carcinogenesis. 1999;20:1737-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 213] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 3. | Zhang HY, Zhang ZY. Pinus massoniana Lamb. Zhongguo Zhongyao Ziyuan Zhiyao. Beijing: Sci-ence Press 1994; 146–147. |
| 4. | Cui YY, Xie H, Wang JF. Broad-spectrum inhibition of Pinus massoniana bark extract (PMBE) to human cancer cells cul-tured in vitro. Yaoxue Jinzhan. 2004;28:418-421. |
| 5. | Rohdewald P. A review of the French maritime pine bark extract (Pycnogenol), a herbal medication with a diverse clinical pharmacology. Int J Clin Pharmacol Ther. 2002;40:158-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 333] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 6. | Shand B, Strey C, Scott R, Morrison Z, Gieseg S. Pilot study on the clinical effects of dietary supplementation with Enzogenol, a flavonoid extract of pine bark and vitamin C. Phytother Res. 2003;17:490-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Huynh HT, Teel RW. Selective induction of apoptosis in human mammary cancer cells (MCF-7) by pycnogenol. Anticancer Res. 2000;20:2417-2420. [PubMed] |
| 8. | Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38285] [Cited by in RCA: 39421] [Article Influence: 938.6] [Reference Citation Analysis (0)] |
| 9. | Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589-601. [PubMed] |
| 10. | Zhu XQ, Wang GS, Zhang XJ, Zhao Q. Effects of sodium ferculate on the viability and apoptosis of human hepatoma BEL-7404 cells. Shijie Huaren Xiaohua Zazhi. 1999;7:715-716. |
| 11. | Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1368] [Cited by in RCA: 1406] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 12. | Yuan J, Gu ZL, Chou WH, Kwok CY. Elemene induces apoptosis and regulates expression of bcl-2 protein in human leukemia K562 cells. Zhongguo Yao Li Xue Bao. 1999;20:103-106. [PubMed] |
| 13. | Kuo ML, Huang TS, Lin JK. Curcumin, an antioxidant and anti-tumor promoter, induces apoptosis in human leukemia cells. Biochim Biophys Acta. 1996;1317:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 263] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 14. | Gautam SC, Xu YX, Pindolia KR, Janakiraman N, Chapman RA. Nonselective inhibition of proliferation of transformed and nontransformed cells by the anticancer agent curcumin (diferuloylmethane). Biochem Pharmacol. 1998;55:1333-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Hockenbery DM. bcl-2 in cancer, development and apoptosis. J Cell Sci Suppl. 1994;18:51-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 2.6] [Reference Citation Analysis (0)] |