Published online Jul 28, 2005. doi: 10.3748/wjg.v11.i28.4390
Revised: May 23, 2004
Accepted: May 5, 2005
Published online: July 28, 2005
AIM: To study whether severe acute respiratory syndrome coronavirus (SARS-CoV) could be excreted from digestive system.
METHODS: Cell culture and semi-nested RT-PCR were used to detect SARS-CoV and its RNA from 21 stool and urine samples, and a kind of electropositive filter media particles was used to concentrate the virus in 10 sewage samples from two hospitals receiving SARS patients in Beijing in China.
RESULTS: It was demonstrated that there was no live SARS-CoV in all samples collected, but the RNA of SARS-CoV could be detected in seven stool samples from SARS patients with any one of the symptoms of fever, malaise, cough, or dyspnea, in 10 sewage samples before disinfection and 3 samples after disinfection from the two hospitals. The RNA could not be detected in urine and stool samples from patients recovered from SARS.
CONCLUSION: Nucleic acid of SARS-CoV can be excreted through the stool of patients into sewage system, and the possibility of SARS-CoV transmitting through digestive system cannot be excluded.
- Citation: Wang XW, Li JS, Guo TK, Zhen B, Kong QX, Yi B, Li Z, Song N, Jin M, Wu XM, Xiao WJ, Zhu XM, Gu CQ, Yin J, Wei W, Yao W, Liu C, Li JF, Ou GR, Wang MN, Fang TY, Wang GJ, Qiu YH, Wu HH, Chao FH, Li JW. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J Gastroenterol 2005; 11(28): 4390-4395
- URL: https://www.wjgnet.com/1007-9327/full/v11/i28/4390.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i28.4390
By the end of 2002, there were reports from Guangdong Province in southern China of cases of severe acute respiratory syndrome (SARS). Over 8 439 SARS cases and 812 SARS-related deaths were reported to WHO from 32 countries around the world till 5th July, 2003[1,2]. In response to this outbreak, WHO coordinated an international collaboration that included clinical, epidemiological, laboratory investigations, and initiated efforts to control the spread of SARS. Attempts to identify the etiology of SARS outbreak were successful during the 3rd wk of March 2003, when laboratories in the USA, Canada, Germany, Hong Kong, and China isolated a novel coronavirus from SARS patients[3-6]. Unlike other human coronaviruses, it was possible to isolate the novel coronavirus in Vero cells. Evidence of the coronavirus infection was documented in SARS patients throughout the world. The coronavirus RNA was frequently detected in respiratory specimens, and convalescent-phase serum specimens from SARS patients containing antibodies that reacted with the coronavirus. There was a strong evidence that this new virus was etiologically linked to the outbreak of SARS[5-8].
Investigations of the global outbreak of SARS have shown that the major mode of transmission of SARS virus was through close personal contact, in particular exposure to droplets of respiratory secretions from an infected person[1,9-14]. While in a cluster of SARS cases in an apartment block in Hong Kong, sewage was believed to have played a role through droplets containing coronavirus from the sewage system[11,12]. However, there is no direct evidence to prove that the coronavirus exists in sewage system and is contagious.
In order to confirm whether the digestive system was a possible major transmission way of SARS-CoV, cell culture and the semi-nested RT-PCR were used to directly detect SARS-CoV and its RNA. A kind of electropositive filter media particle[15] was used to concentrate the SARS-CoV from the sewage of hospitals receiving SARS patients in Beijing of China, and then the virus and its RNA were detected.
To identify viruses that existed in stools, urine samples, and sewage system, we inoculated a variety of specimens onto Vero E6. Because of the toxicity of sewage concentrates, all cell cultures were inoculated in the presence of growth medium for 1 h at 37 °C. This procedure virtually eliminated problems with the toxicity of sewage concentrates. Medium was replaced after 1-2 d of incubation. Culture was terminated 7 d after inoculation, and the culture was observed daily for cytopathic effects. Cultures exhibiting identifiable cytopathic effects were subjected to several procedures to identify the cause of the effect[16-18]. If there was no cytopathic effect on the cell culture, the supernate was harvested and added into additional flasks to isolate viruses. The cultures were then used until three generations without cytopathic effects.
Twenty-one stool and urine samples were collected from the Xiao Tang Shan Hospital and 309 Hospital of PLA, which were specially assigned to receive SARS patients in Beijing in 2003, among which 11 samples were collected from the SARS patients with any one of the symptoms of fever, malaise, cough, or dyspnea, and 10 samples were from recovered patients.
Ten sewage samples were collected at 7 o'clock in the morning from Xiao Tang Shan Hospital and 309 Hospital of PLA for 7 d. Two thousand and five hundred milliliters of sewage before disinfection or 25 000-50 000 mL after disinfection by chlorine was collected.
The positively charged filter media particles, which were used to concentrate SARS-CoV from sewage, were prepared as previously described[15].
The residual chlorine in sewage was determined by the N, N, diethyl-p-phenyldiamine colorimetric method[19].
Two thousand and five hundred milliliters and 25 000 mL `sewage from the hospitals before or after disinfection by chlorine were placed in a 25-L capacity plastic bucket, and 10 mL Na2S2O3 (100 g/L) was added to neutralize the residual chlorine. Five hundred or 820 g filter media was packed in a polymethyl methacrylate column (89 or 130 mm i.d.). The filter media bed height was 14 cm. The flow rate was kept at 10 mL/min per cm2 of the filter surface area. The adsorbed viruses were eluted from filter media with 700 or 900 mL 6 nutrient broth (pH 7.2). The collected eluates were reconc-entrated by PEG precipitation and centrifugation. The pellets were resuspended in 40 mL PBS and assayed.
Virus RNA extracting kit (TRIzol Reagent) made by InvitrogenTM Life Technologies for extraction of exceedingly pure viral RNA was utilized in our experiment to extract virus RNA, and all procedures were strictly implemented in accordance with the reagent instruction manual.
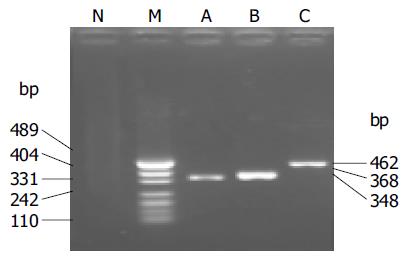
Three sets of primers from WHO Network Laboratories[20] were used to detect the SARS-CoV RNA: Cor-p-F2 (+) 5'-CTAACATGCTTAGGATAATGG-3', Cor-p-F3 (+) 5'-GCCTCTCTTGTTCTTGCTCGC-3', and Cor-p-R1 (-) 5-CAGGTAAGCGTAAAACTCATC-3. Cor-p-F2/Cor-p-R1 gave a 368-bp product, and Cor-p-F3/Cor-p-R1 yielded a 348-bp segment.
A pair of consensus primers of enteroviruses was from the 5 non-coding region because of their presence in many enterovirus serotypes. The sequences of primers were as follows: E1 5-ATTGTCACCATAAGCAGCCA-3, E2 5-CCAGCACTTCTGTTTCCCCGG-3, and the product size was 440 bp[21].
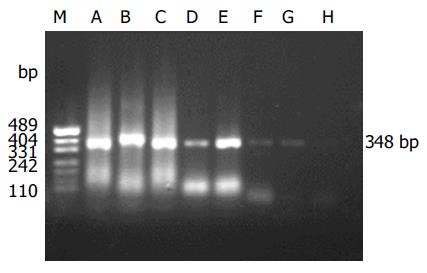
Two microliters of RNA solution was analyzed with RT-PCR assay. The KaTaRa one step RNA PCR kit (KaTaRa Biotechnology, Dalian) was used for the reaction (20 mL total volume). Positive and negative RT-PCR controls were included in each run. Reactions contained 10 mL of buffer concentrate, 2 mmol/L of magnesium sulfate, 0.8 mL of enzyme mixture, and 1.9 mmol/L of each of Cor-p-F2 and Cor-p-R1 primers. Thermal cycling comprised 42 °C for 30 min, 95 °C for 3 min; 10 cycles of 95 °C for 10 s, 55 °C for 15 s (decreasing by 1 °C per cycle), 72 °C for 40 s; 40 cycles of 95 °C for 10 s, 56 °C for 10 s, and 72 °C for 40 s. To confirm the PCR products, a semi-nested PCR was developed. The template was the first PCR product, and the primers were Cor-p-F3 and Cor-p-R1, and yielded a 348-bp product. The amplification efficiency of the primers was confirmed by SARS-CoV of BJ-01 isolated from a SARS patient in Guangzhou city, China (Figure 2).
As most of enteroviruses can also grow on the Vero cells, and yield cytopathic effects, enteroviruses should be detected by the specific primers[21]. The RT-PCR method was similar to that for SARS-CoV.
PCR products were analyzed by electrophoresis with 15 g/L agarose gels containing 0.5 mg of ethidium bromide per mL, and visualized with UV illumination and photographed. DNA molecular size standards (100-bp ladder, Gibco/BRL) were included in each run of agarose gel electrophoresis.
In view of the serious nature of SARS and the person-to-person transmission, all clinical specimens were treated in a biosafety level 3 environment. All divisions into aliquots, pipetting, concentration for small sewage and culture attempts were performed in laminar-flow safety cabinets. A similar environment was used when specimens from which nucleic acid was to be extracted and placed in a buffer solution.
The PCR products from four different samples were purified with the QIAquick PCR purification kit (QIANEN, Inc.) and sequenced with the ABI PRISM dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystem) following the manufacturer's instructions. The sequences were compared with the genome of SARS-CoV in the GenBank and EMBL databases by using the FASTA program of the GCG.
The detection specificity and sensitivity of semi-nested RT-PCR were confirmed by the isolated SARS-CoV (BJ-01) from Institute of Microbiology and Epidemiology, Academy of Military Medical Sciences, Beijing, China. It was shown that two amplicons were yielded, which were in agreement with the information on the designed primers (Figure 1). The minimum amount of SARS-CoV RNA detected by semi-nested PCR was equivalent to 10 TCID50 (Figure 2).
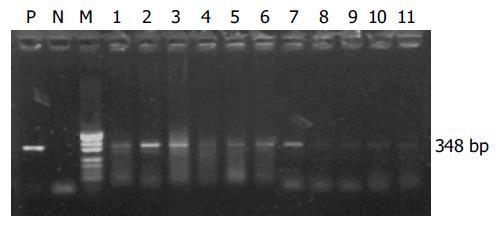
All the 21 stool samples tested for the presence of infectious SARS-CoV in cell culture were negative. SARS-CoV RNA could be detected in 7 of 11 stool samples from patients with symptoms by semi-nested RT-PCR (Figure 3). However, SARS-CoV RNA could not be detected from the samples of patients who recovered.
All the 21 urine samples tested for the presence of infectious SARS-CoV in cell culture were also negative. SARS-CoV RNA could not be detected from the samples or the supernate of cell cultures by semi-nested RT-PCR.
All sewage samples tested for the presence of infectious SARS-CoV in cell culture were negative. SARS-CoV RNA could be found in the concentrates of sewage from the two hospitals by semi-nested PCR, and in the inoculated cells of the sewage concentrates from 309 Hospital but not from Xiao Tang Shan Hospital. However, SARS-CoV RNA copies in the samples were too low to be detected by the first amplification reaction, the semi-nested RT-PCR in which the products of first amplification reaction were the template of the second PCR, gave the positive amplification results (Tables 1 and 2).
| Date | Cell cult | Concentrate +PCR | Inoculated cells+PCR | Entero. PCR |
| 11 June | - | + | - | - |
| 12 June | - | + | - | - |
| 13 June | - | + | - | - |
| 14 June | - | + | - | - |
| 15 June | - | + | - | - |
| 16 June | - | + | - | - |
The samples (25 000 or 50 000 mL ) from the two hospitals were all negative by the infectivity methods. SARS-CoV RNA was detected from the concentrates and inoculated cells in three samples (June 11, 13, and 15) from 309 Hospital by semi-nested RT-PCR, while the other samples were negative (Tables 3 and 4).
| Date | Cell culture | Concentrate +PCR | Inoculated cells+PCR | Enterovirus PCR |
| 11 June | - | - | - | - |
| 12 June | - | - | - | - |
| 13 June | - | - | - | - |
| 14 June | - | - | - | - |
| 15 June | - | - | - | - |
| Date | Cell culture | Concentrate +PCR | Inoculated cells+PCR | Enterovirus PCR |
| 11 June | - | + | + | - |
| 12 June | - | - | - | - |
| 13 June | - | + | + | - |
| 14 June | - | - | - | - |
| 15 June | - | + | + | - |
The PCR products from the sewage samples of the two hospitals were sequenced, and submitted to GenBank. The accession numbers are bankit579728 and bankit579738, respectively. Comparison of the nucleotide sequences of PCR products with data from GenBank revealed that the sequences of the PCR products were close to those of SARS-COV genomes, showing about 99% nucleotide homolog.
Most SARS cases to date have occurred in young adults. The health care workers in hospitals, patient family members and international travelers were the commonly infected people[22,23].
The isolation of a novel coronavirus was obtained from the respiratory secretions of patients with SARS, and points to the etiologic association with SARS[3-6,8,24-26].
The mechanism of transmission of SARS-CoV is not yet understood completely. However, the fact that transmission has been limited to close contacts with patients, such as household members, health care workers, or other patients who were not protected with contact or respiratory precautions, suggests that either droplet secretions or direct or indirect contact probably has a role[1,10-14,27].
On 15th April, 2003, health authorities reported a total of 321 individuals infected with SARS virus who were residents in Amoy Gardens. A large proportion of cases were concentrated in vertically linked flats in a single building, Block E. On April 17, the Hong Kong Government announced that not one single factor could account for the outbreak in Block E of Amoy Gardens, and attention was focused on possible transmission via the sewage system because laboratory studies showed that patients with the disease excreted coronaviruses in their stools and these viruses were able to survive much longer in feces than on ordinary surfaces, and noted a swab sample taken from the toilet of an infected resident showed a positive test for the coronavirus genetic material, and about 60% of patients in Amoy Gardens had diarrhea during their illness, and probably would have discharged a large amount of viruses into the soil stacks. Finally, the virus would spread with water droplets through the U-traps of the floor drains, which were dried up in many cases[11,24,28,29].
The elevated levels of aspartate aminotransferase and lactate dehydrogenase indeed suggest that SARS-CoV was also replicating outside the respiratory tract[1]. Electron micro-scopic examination showed that virus-like particles with 100-150 nm in diameter were found in cytoplasm and dilated reticular endoplasm of the infected alveolar epithelial cells and endothelial cells[6,26,30,31]. Shedding of the virus in feces might be an additional source of spreading, provided the virus was stable in this environment[8].
Tsang et al[1] reported that there were three nurses who worked at Hospital B, where a patient was admitted and remained for 6 d for treatment of pneumonia before he was transferred to Hospital C. During this period, the nurses spent five 8-h shifts stationed on the general ward where the patient was hospitalized. The three nurses recalled close encounter with the patient during which they cleaned him when he had fecal incontinence after an episode of diarrhea on March 3, 2003. The nurses did not wear masks or gowns during their routine nursing care of any patients on the ward and finally were all infected.
The detection of SARS-CoV in fecal and serum samples from patients, as well as in respiratory specimens, suggested that this virus, like many animal coronaviruses, might spread both by fecal contamination and by respiratory droplets[4].
Zhang reviewed the data of SARS transmission and believed that, as previously described, most coronaviruses could cause either a respiratory or an enteric disease, which is also transmitted by the fecal-oral route. During this outbreak of SARS, symptoms of the gastrointestinal tract in patients were noticed. Many investigators[1,10,32] found that gastroi-ntestinal symptoms, including diarrhea (19-50%), nausea and vomiting (19.6%), and abdominal pain (13%) were common in SARS patients.
All the above reports suggested that stools of SARS patients or sewage containing stools of SARS patients would transmit the coronavirus. However, except that the positive PCR results were obtained in some patient stools, there were no reports that live viruses were present in patient stool or sewage.
In this study, we isolated and detected the SARS-CoV in stools, urine samples, and sewage from hospitals which were assigned specially to receive SARS patients in Beijing of China. Just as expected, SARS-CoV RNA was detected from stools of patients, but no live viruses were isolated from stool samples, and no SARS-CoV RNA was found in all stools from patients who recovered. It is suggested that the nucleic acid of SARS-CoV could be really excreted from stools of patients, but infectious SARS-CoV could not be confirmed to excrete through the digestive system. No live virus and its RNA were isolated from the urine samples of patients. It is suggested that SARS-CoV and its RNA could not be excreted from the urinary system.
In order to explore the growth and decline of SARS-CoV and its RNA in environment, SARS-CoV and its RNA were isol-ated from sewage of hospitals. Although the concen-tration method of SARS-CoV from sewage has not been reported yet, the concentration of enteroviruses from water using different methods was reported, and the electropositive filters have been considered as the most promising method[34,35].
We developed a simple method for concentration of entero-viruses from water with electropositive particles, the adsorption of bacteriophage f2 was reliable and efficient, not affected by the pH value, temperature, turbidity, and organic materials in water, and gave a recovery of 88.7% for poliovirus I and a comparable recovery of HAV, CoxB3, and Echo 7 from 100 L of tap water[15].
We attempted to concentrate SARS-CoV in sewage from Xiao Tang Shan Hospital and 309 Hospital in Beijing by the electropositive particle adsorption method. The sewage systems in these two hospitals were similar, i.e. the sewage was collected from each isolation ward and converged into the reaction sedimentation basin, and disinfectant (chlorine) was added to inactivate SARS-CoV and other pathogenic microorganisms; finally, the sewage was discharged from the reaction sedimentation basin after a 60-min reaction.
Results of testing for the presence of SARS-CoV in the sewage indicated that no infectious SARS-CoV or live virus could be recovered in these two hospitals. The nucleic acid of SARS-CoV was found in the sewage before disinfection from both hospitals by semi-nested RT-PCR, while after disinfection of sewage by chlorine, SARS-CoV RNA could only be detected in the samples taken on 11th, 13th, and 15th June, 2003 from 309 Hospital.
Cell culture is a very demanding test. However, negative cell culture results or RT-PCR results could not exclude the presence of SARS-CoV. The detection of SARS-CoV from SARS patients could be negative for the following reasons[17]. Patients were not infected with SARS coronavirus, the illness was due to another infectious agent (virus, bacterium, fungus) or a non-infectious cause. The test results were incorrect. Current tests need to be further developed to improve their sensitivity. SARS-CoV was so susceptible to environments that it was inactivated quickly out of the body. SARS-CoV might have been inactivated or eliminated by immunoglobulins (antibody) from the recovered patients before excretion, or in the sewage. Palmer et al[35] reported that human imm-unoglobulins were used to eliminate the enteroviruses in concentrated sewage when they evaluated the immunodeficiency virus (HIV) in sewage effluent by infectivity assay and RT-PCR.
Hong Kong Government explained the reasons of a cluster of SARS cases in Amoy Gardens and believed that there was a combination of factors, including the presence of an index patient who caused the first batch of infections, person-to-person spread, transmission via the sewage system, and environmental contamination[11,29]. This study demons-trated that SARS-CoV RNA could be excreted through the feces or/and urine samples of patients into sewage system.
In conclusion, this study demonstrated that there was SARS-CoV RNA in stool samples of patients with symptoms and in sewage of hospitals though there was no live SARS-CoV isolated from all samples. It provides evidence that the nucleic acid of SARS-CoV can be excreted through the stools of patients into sewage system, but cannot exclude the possibility of SARS-CoV transmitting through the digestive system. Much attention should be paid to the treat-ment of stools of patients and the sewage of hospitals receiving SARS patients.
The authors thank Dr. Da-Sheng Zhao, De-Xue Li, Jian-Zhong Sun, Zhong-Hou Huo and Yun-Bo Li from the P3 Laboratory Center for Microorganism Detection, China for supporting of this project; Dr. Fu-Yu Wang, Ying-Kai Li, Meng-Fu Zhu, Jian-Yong Su, Cheng-Yuan Gong, Wu-Chun Chao, Tai-Thi Gong, Bing-Yin Si and Bao-Zhong Guo for providing many reagents, helpful guidance and discussion; Drs. Hong-Wei Zhao, Xin-An Du, Zong-Ze Wang, Ling-Jia Qian, Qing-Yu Zhu, Xiao-Jun Zhang, Tao-Xing Shi, Fei Yu, Jian-Zhong Man, Fan-Rong Zeng, Bang-Rong Han, Yue Jiang, Zhu-Ge Xi, Zhi-Peng Ju and Hua-Shan Zhang for advice and organizing the experiments; the Center for Logistics, Xiao-Tang Shan Hospital for technical support and cooperation. We are also indebted to Professor Su-Qi Cheng for English revision.
Science Editor Zhang JZ and Wang XL Language Editor Elsevier HK
| 1. | Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, Chan-Yeung M, Lam WK, Seto WH, Yam LY, Cheung TM. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977-1985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 718] [Cited by in RCA: 727] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 2. | World Health Organization. SARS: breaking the chains of transmission. Available from: http: //www.who.int/features/2003/07/en. |
| 3. | Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen MH. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1864] [Cited by in RCA: 1882] [Article Influence: 85.5] [Reference Citation Analysis (0)] |
| 4. | Holmes KV. SARS-associated coronavirus. N Engl J Med. 2003;348:1948-1951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 206] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stöhr K, Osterhaus AD. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 638] [Cited by in RCA: 621] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 6. | Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3147] [Cited by in RCA: 3072] [Article Influence: 139.6] [Reference Citation Analysis (0)] |
| 7. | Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1538] [Cited by in RCA: 1559] [Article Influence: 70.9] [Reference Citation Analysis (0)] |
| 8. | Qin ED, Zhu QY, Peng WM, Jiang T, Fan BC, Chang GH, Yu M, Si BY, Liu BH, Deng YQ. Determination of the partial polymerase gene sequence of novel coronavirus isolated from lung tissue of SARS patients. Junshi Yixue Kexueyuan Yuankan. 2003;27:81-83. |
| 9. | Enserink M, Vogel G. Infectious diseases. Hungry for details, scientists zoom in on SARS genomes. Science. 2003;300:715-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1563] [Cited by in RCA: 1602] [Article Influence: 72.8] [Reference Citation Analysis (1)] |
| 11. | World Health Organization. Amoy Gardens investigation findings make public. Available from: http: //www.who.int/csr/sars/en (17 April,2003). |
| 12. | Cyranoski D, Abbott A. Apartment complex holds clues to pandemic potential of SARS. Nature. 2003;423:3-4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M. Identification of severe acute respiratory syndrome in Canada. N Engl J Med. 2003;348:1995-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 786] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 14. | Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu-Raddad LJ, Ho LM, Thach TQ, Chau P. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761-1766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 638] [Cited by in RCA: 617] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 15. | Li JW, Wang XW, Rui QY, Song N, Zhang FG, Ou YC, Chao FH. A new and simple method for concentration of enteric viruses from water. J Virol Methods. 1998;74:99-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 32] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | World Health Organization. Recommendations for laboratories testing by PCR for presence of SARS coronavirus-RNA. Available from: http: //www.who.int/csr/sars/coronarecommendation/en. |
| 17. | World Health Organization. Severe Acute Respiratory Syndrome (SARS): Laboratory diagnostic tests. Available from: http: //www.who.int/csr/sars/diagnostictests/en. |
| 18. | World Health Organization. Use of laboratory methods for SARS diagnosis. Available from: http: //www.who.int/csr/sars/labmethods/en. |
| 19. | Olivieri VP, Snead MC, Krusé CW, Kawata K. Stability and effectiveness of chlorine disinfectants in water distribution systems. Environ Health Perspect. 1986;69:15-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 15] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | World Health Organization. PCR primers for SARS developed by WHO Network Laboratories. Available from: http: //www.who.int/csr/sars/primers/en. |
| 21. | Zoll GJ, Melchers WJ, Kopecka H, Jambroes G, van der Poel HJ, Galama JM. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J Clin Microbiol. 1992;30:160-165. [PubMed] |
| 22. | World Health Organization. Update 58 - First global consultation on SARS epidemiology, travel recommendations for Hebei Province (China), situation in Singapore. Available from: http: //www.who.int/csr/sars/archive/2003_05_17/en/. |
| 23. | World Health Organization. SARS epidemiology to date. Available from: http: //www.who.int/csr/sars/epi2003_04_11/en/. |
| 24. | Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2097] [Cited by in RCA: 2154] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 25. | World Health Organization. Update 31 - Coronavirus never before seen in humans is the cause of SARS. Available from: http: //www.who.int/csr/sars/archive/2003_04_16/en. |
| 26. | Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3277] [Cited by in RCA: 3315] [Article Influence: 150.7] [Reference Citation Analysis (0)] |
| 27. | Seto WH, Tsang D, Yung RW, Ching TY, Ng TK, Ho M, Ho LM, Peiris JS. Effectiveness of precautions against droplets and contact in prevention of nosocomial transmission of severe acute respiratory syndrome (SARS). Lancet. 2003;361:1519-1520. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 568] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 28. | World Health Organization. Update 32-Situation in China and Hong Kong, status of diagnostic tests. Available from: http: //www.who.int/csr/sarsarchive/2003_04_17/en. |
| 29. | Available from: http: //www.info.gov/csr/sars/labmethods/en. |
| 30. | Wang CE, Qin ED, Gan YH, Li YC, Wu XH, Cao JT, Yu M, Si BY, Yan G, Li JF. Pathological observation on sucking mice and Vero E6 cells inoculated with SARS samples. Jiefangjun Yixue Zazhi. 2003;28:383-384. |
| 31. | Hong T, Wang JW, Sun YL, Duan SM, Chen LB, Qu JG, Ni AP, Liang GD, Ren LL, Yang RQ. Chlamydia-like and coronavirus-like agents found in dead cases of atypical pneumonia by electron microscopy. Zhonghua YiXue ZaZhi. 2003;83:632-636. [PubMed] |
| 32. | Riley S, Fraser C, Donnelly CA, Ghani AC, Abu-Raddad LJ, Hedley AJ, Leung GM, Ho LM, Lam TH, Thach TQ. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 654] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 33. | Sobsey MD, Jones BL. Concentration of poliovirus from tap water using positively charged microporous filters. Appl Environ Microbiol. 1979;37:588-595. [PubMed] |
| 34. | Sobsey MD, Glass JS. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl Environ Microbiol. 1980;40:201-210. [PubMed] |
| 35. | Palmer CJ, Lee MH, Bonilla GF, Javier BJ, Siwak EB, Tsai YL. Analysis of sewage effluent for human immunodeficiency virus (HIV) using infectivity assay and reverse transcriptase polymerase chain reaction. Can J Microbiol. 1995;41:809-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |