Published online Jul 21, 2005. doi: 10.3748/wjg.v11.i27.4268
Revised: November 23, 2004
Accepted: November 26, 2004
Published online: July 21, 2005
AIM: To identify the distribution of N-acetyltrasferase 2 (NAT2) polymorphism in Hebei Han Chinese and the effects of the polymorphism on the development of colorectal cancer.
METHODS: We performed a hospital-based case-control study of 237 healthy individuals and 83 colorectal cancer patients of Hebei Han Chinese. DNA was extracted from peripheral blood and cancer tissues. The genotypes of the polymorphisms were assessed by PCR-restriction fragment length polymorphism(RFLP).
RESULTS: There were four NAT2 alleles of WT, M1, M2, and M3 both in the healthy subjects and in the patients, and 10 genotypes of WT/WT, WT/M1, WT/M2, WT/M3, M1/M1, M1/M2, M1/M3, M2/M2, M2/M3, M3/M3. M2 allele was present in 15.61% of healthy subjects and 29.52% of patients (χ2 = 15.31, P < 0.0001), and M3 allele was present in 30.59% of healthy subjects and 16.87% of patients (χ2 = 25.33, P < 0.0001). There were more WT/M2 (χ2 = 34.42, P < 0.0001, odd ratio = 4.99, 95%CI = 2.27-9.38) and less WT/M3 (χ2 = 3.80, P = 0.03) in the patients than in the healthy subjects. In 70.3% of the patients, there was a difference in NAT2 genotype between their tumors and blood cells. Patients had more WT/M2 (χ2 = 5.11, P = 0.02) and less M2/M3 (χ2 = 4.27, P = 0.039) in their blood cells than in the tumors. Furthermore, 53.8% (7/13) of M2/M3 in tumors were from WT/M2 of blood cells.
CONCLUSION: There is a possible relationship between the NAT2 polymorphisms and colorectal cancer in Hebei Han Chinese. The genotype WT/M2 may be a risk factor for colorectal cancer.
-
Citation: He LJ, Yu YM, Qiao F, Liu JS, Sun XF, Jiang LL. Genetic polymorphisms of
N -acetyltransferase 2 and colorectal cancer risk. World J Gastroenterol 2005; 11(27): 4268-4271 - URL: https://www.wjgnet.com/1007-9327/full/v11/i27/4268.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i27.4268
Colorectal cancer is one of the common tumors in Hebei Province of China. N-acetyltransferase 2 (NAT2) is polymorphic and catalyzes both N-acetylation (usually deactivation) and O-acetylation (usually activation) of a variety of heterocyclic amine drugs and carcinogens[1]. Heterocyclic amines which are found mainly in well-cooked meat, require metabolic activation to function as mutagens and animal carcinogens. Enzymes such as cytochrome P4501A2 (CYP1A2) and NAT2 perform this task. Genetic studies in humans have shown that individuals may be classified as rapid or slow acetylators according to the rates at which drugs are acetylated by NAT2[2]. NAT2 polymorphism is a representative genetic trait of individual’s susceptibility to several cancers[3]. Epidemiological studies suggest that the NAT2 acetylation polymorphisms modify the risk of developing carcinoma of urinary bladder, colorectal and breast cancer, head and neck cancer, pulmonary and prostatic cancer. Associations between slow NAT2 acetylator genotypes and urinary bladder cancer as well as between rapid NAT2 acetylator genotypes and colorectal cancer are the most consistently reported[3]. However, whether there is a true association between colorectal cancer and NAT2 polymorphism is controversial. Human epidemiological studies suggest that rapid acetylator phenotypes may be associated with higher incidences of colorectal cancer in American populations[4]. Researchers in UK[5] found that no significant differences in NAT2 allelic frequencies (WT, M1, M2, M3 alleles) or genotypes are observed between colorectal cancer patients and the healthy population. Ethnic differences exist in NAT2 genotype frequencies that may be a factor in cancer incidence, emphasizing the need to investigate the distribution frequency of NAT2 genotypes and the association of their polymorphism with colorectal cancer in Hebei Han Chinese population.
We identified the distribution of NAT2 polymorphisms in Chinese healthy individuals and colorectal cancer patients from Hebei Province. We also performed a case-control study to investigate whether the NAT2 genetic polymorphism was a risk factor for colorectal cancer in these Chinese population.
We conducted a case-control study of colorectal cancer patients between April 2001 and April 2003. Both patients and controls were Hebei Han Chinese. Eighty-three patients (46 males, 37 females) with histopathologically confirmed diagnosis of colorectal cancer, were recruited from the Department of Surgery at Hebei No. 4 Hospital. Their mean age was 47.5 years (ranging 25-77 years). The control group consisted of 237 (124 males, 113 females) healthy individuals selected from the blood donors in Hebei Province Blood Center. Their average age was 42.6 years (ranging 20-80 years).
The peripheral blood was treated with EDTA. Tumor samples were obtained from colorectal cancer patients and immediately frozen in liquid nitrogen and stored at -80°C until used. Genomic DNA was isolated from peripheral leukocytes by PEL-FREEZE reagent protocol or from the tumor samples by the standard procedure of proteinase K/RNase digestion and phenol/chloroform extraction.
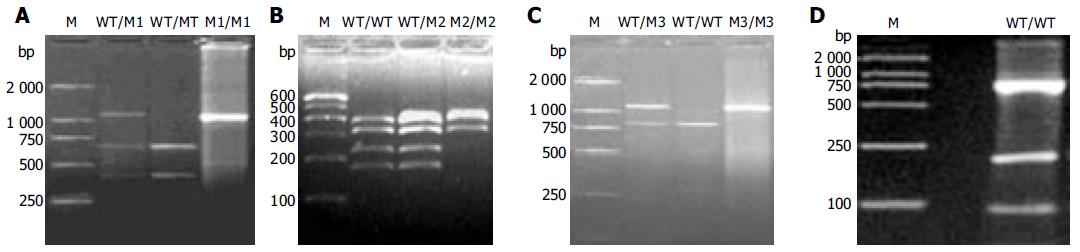
PCR-RFLP was performed to investigate the NAT2 allele. PCR was performed in 40 L reaction mixture containing 50-200 ng DNA, 20 mmol/L of each primer, 1.25 mmol/L of dNTPs, 25 mmol/L of MgCl2, 1´PCR buffer, 5U of thermostable Taq DNA polymerase using a programmable thermocycler. The primer sequences were 5’-GGA ACA AAT TGG ACT TGG-3’ and 5’-TCT AGC ATG AAT CAC TCT GC-3’. PCR conditions were 5 min at 95°C; 35 cycles of 1 min at 95°C, 1 min at 57°C and 1.5 min at 72°C; followed by a final extension for 5 min at 72°C. After amplification, the 1 093 bp PCR-products were digested with KpnI (M1 allele), BamHI (M3 allele), MspI/AluI (M4 allele)for 16-18 h at 37°C and TaqI (M2 allele) for 16-18 h at 65°C separately in a 25 L final volume. Digested fragments were then separated by electrophoresis on 20 g/L agarose gel (M1, M3) or 40 g/L agarose gel (M2, M4) and visualized by ethidium bromide staining. All the experiments included positive and negative controls for each studied polymorphism.
Statistical analysis was performed with SAS 6.0 for Windows. Hardy-Weinberg equilibrium test was carried out using statistical software for linkage analysis. χ2 and Fisher’s exact test were used for comparisons of frequencies. Associations were expressed as odd ratios (OR) with 95% confidence interval (95%CI). P < 0.05 was considered statistically significant.
The results of genotype analysis are shown in Figure 1. In 237 healthy and 83 colorectal cancer patients of Hebei Han Chinese, there were four kinds of NAT2 alleles including wild-type gene WT allele and polymorphisms of M1, M2, and M3 alleles (Table 1). No M4 polymorphism was found. Ten genotypes including WT/WT, WT/M1, WT/M2, WT/M3, M1/M1, M1/M2, M1/M3, M2/M2, M2/M3, M3/M3 and their frequencies in both groups are shown in Table 2. M3 (59.67%) was the major polymorphism in healthy group. The distribution of NAT2 genotypes in healthy group was consistent with Hardy-Weinberg equilibrium (χ2 = 7.2533, g = 8, P > 0.5). In healthy group, 71.3% and 28.7% were classified as rapid-acetylator genotypes (WT/WT, WT/Mx, x = 1, 2, 3) and slow-acetylator genotypes (Mx/Mx), respectively. The frequency of WT/M3 (29.1%) in the rapid-acetylator genotypes was the highest.
| Rapid-acetylator genotype | Slow-acetylator genotype | ||||||||||||
| WT/WT | WT/M1 | WT/M2 | WT/M3 | Sum | M1/M1 | M1/M2 | M1/M3 | M2/M2 | M2/M3 | M3/M3 | Sum | ||
| Controls | n | 62 | 9 | 29 | 69 | 169 | 1 | 4 | 9 | 5 | 31 | 18 | 68 |
| % | 26.2 | 3.8 | 12.2 | 29.1 | 71.3 | 0.4 | 1.7 | 3.8 | 2.1 | 13.1 | 7.6 | 28.7 | |
| Patients | n | 15 | 1 | 35 | 14 | 65 | 1 | 2 | 4 | 3 | 6 | 2 | 18 |
| % | 18.1 | 1.2 | 42.2 | 16.9 | 78.3 | 1.2 | 2.4 | 4.8 | 3.6 | 7.2 | 2.4 | 21.7 | |
| χ2 | 2.2 | 0.64 | 34.4 | 3.8 | 1.54 | 0 | 0 | 0.007 | 0.12 | 2.06 | 2.01 | 1.54 | |
| P | 0.14 | 0.42 | < 0.0001 | 0.03 | 0.22 | 1 | 1 | 0.94 | 0.73 | 0.15 | 0.16 | 0.22 | |
The frequency of allele M2 (29.5%) was the highest in the 83 patients, 18 (21.7%) were slow-acetylator genotypes (Mx/Mx), and 65 (78.3%) were rapid-acetylator genotypes (WT/WT, WT/Mx), 35 (53.8%) of them being WT/M2 (Table 2).
M3 allele was present in 30.59% (154/474) of the healthy subjects and in 16.87% (28/166) of the patients (χ2 = 25.33, P < 0.0001). The M2 allele frequency was significantly different between healthy (15.61%) and patient groups (29.52%) (χ2 = 15.31, P < 0.0001). There was no difference in rapid-acetylator or slow-acetylator frequency between healthy and patient groups. However, there was a significant difference between WT/M2 (χ2 = 34.42, P < 0.0001) and WT/M3 (χ2 = 3.80, P < 0.05) compared to the healthy controls. The risk associated with combinations of NAT2 genotypes is shown in Table 3. The genotype WT/M2 might be a risk factor for colorectal cancer (OR = 4.99, 95%CI = 2.27-9.38).
| Genotypes | Control | Patients | OR (95%CI) | ||
| n | % | n | % | ||
| WT/WT | 62 | 26.2 | 15 | 18.1 | 1 |
| WT/M2 | 29 | 12.2 | 35 | 42.2 | 4.99 (2.27 - 9.38) |
| WT/M1 | 9 | 3.8 | 1 | 1.2 | 0.46 (0.05 - 3.92) |
| WT/M3 | 69 | 29.1 | 14 | 16.9 | 0.84 (0.38 - 1.88) |
| WT/WT and WT/Mx | 169 | 71.3 | 65 | 78.3 | 1.59 (0.85 - 2.99) |
| Mx/Mx | 68 | 28.7 | 18 | 21.7 | 1.09 (0.51 - 2.35) |
In 70.4% of patients, there was a difference in NAT2 genotype between tumor tissues and blood cells. The frequency of NAT2 genotypes, especially WT/M2 and M2/M3, was significantly different between tumor tissues and blood cells (χ2 = 5.11, P < 0.05; χ2 = 4.27, P < 0.05, Table 4). The frequency of genotype M2/M3 was 9.3% in peripheral blood cells and 24.1% in cancer tissue. Furthermore, 53.8% (7/13) of the tumors had M2/M3 while the corresponding blood cells had WT/M2.
| WT/M2 | M2/M3 | Others | ||||
| n | % | n | % | n | % | |
| Blood cells | 23 | 42.6 | 5 | 9.3 | 26 | 48.2 |
| Cancer tissues | 12 | 22.21 | 13 | 24.12 | 29 | 53.7 |
Human NAT2 is a genetic polymorphism. Different genotypes (WT/WT, WT/Mx, Mx/Mx, x = 1, 2, 3, 4) are composed of different NAT2 alleles (wild type allele WT, mutation allele Mx). According to the intensity of acetyl action, a different genotype shows a different phenotype (rapid-acetylator genotype and slow-acetylator genotype). Genotypes composed of any two mutation alleles (Mx/Mx) show slow-acetylator genotypes, while those consisted of WT/WT or WT/Mx show rapid-acetylator genotypes. Bell et al[6], reported that there are significant differences in the frequency of WT, M1, and M4 alleles between Caucasian and African-Americans, while the frequency of M2 and M3 alleles is similar. The M4 allele is not detectable in Cancasian-Americans, while 18% African-Americans carry at least one M4 allele. The frequencies of NAT2 WT, M1, M2, M3, and M4 alleles in Singapore Chinese are 0.51, 0.07, 0.32, 0.10, and 0.00, respectively[7]. The proportions of rapid and slow acetylators do also vary remarkably between ethnic groups as well as between populations of different geographical origins[8]. For example, the percentage of slow acetylators is 5% in Canadian Eskimos, over 80% in Egyptians and 90% in Moroccans[9,10]. Singapore Chinese have 72% of rapid-acetylator genotypes. In our study, four NAT2 alleles (WT, M1, M2, M3) and ten NAT2 genotypes (WT/WT, WT/M1, WT/M2, WT/M3, M1/M1, M1/M2, M1/M3, M2/M2, M2/M3, M3/M3) were found in Hebei Han Chinese. M4 was not present in Hebei Han Chinese, while 29.1% of healthy controls carried at least one M3 allele. Hebei Han Chinese had 71.3% of rapid-acetylator genotypes. The frequencies of the NAT2 M4 allele and rapid-acetylator genotypes are similar in both Hebei Han Chinese and Singapore Chinese, but there are significant differences in the frequencies of WT, M1, M2, and M3 between the two populations.
NAT2 is an important enzyme for the detoxification and/or bioactivation of several carcinogenic arylamines, its activity directly affects metabolic response of carcinogen in vivo and intensity of toxic reaction[11]. It was reported that the polymorphism of NAT2 gene is associated with the occurrence of colorectal cancer[12]. Genetic epidemiological studies suggest that rapid acetylator phenotype may be associated with the higher incidence of colorectal cancer in the UK populations[12]. Lee et al[7], also showed that the rapid acetylator genotype is associated with cancer occurring in Singapore Chinese. However, our results suggest that rapid-acetylator genotype or slow-acetylator genotype does not appear to be associated with colorectal cancer development, but there is an association between the WT/M2 genotype and colorectal cancer. The M2 allele was more frequent in the patients than in the healthy subjects (χ2 = 15.31, P < 0.0001). Odds ratio (OR) for genotype WT/M2 was 4.99 (95%CI = 2.27-9.38). The data suggest that genotype WT/M2 may be a risk factor for colorectal cancer in Hebei Han Chinese.
Schnakenberg et al[13], detected the loss of heterozygosity in slow and rapid acetylator genotypes in bladder cancer patients. They observed allelic loss at the NAT2 locus in 11 of 60 tumor samples (18.3%), but not in controls. The present study also demonstrated changes of NAT2 genotypes in 70.4% of the tumor specimens. The rising M2/M3 in tumors seems to be transformed from genotype WT/M2 of blood cells, and also suggests that WT/M2 may be a risk factor for colorectal cancer. Our findings may contribute to the early diagnosis of colorectal cancer.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Pande JN, Pande A, Singh SP. Acetylator status, drug metabolism and disease. Natl Med J India. 2003;16:24-26. [PubMed] |
| 2. | Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506-507:65-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 334] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 3. | Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29-42. [PubMed] |
| 4. | Rodriguez JW, Kirlin WG, Ferguson RJ, Doll MA, Gray K, Rustan TD, Lee ME, Kemp K, Urso P, Hein DW. Human acetylator genotype: relationship to colorectal cancer incidence and arylamine N-acetyltransferase expression in colon cytosol. Arch Toxicol. 1993;67:445-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Sachse C, Smith G, Wilkie MJ, Barrett JH, Waxman R, Sullivan F, Forman D, Bishop DT, Wolf CR. A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis. 2002;23:1839-1849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 195] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | Bell DA, Taylor JA, Butler MA, Stephens EA, Wiest J, Brubaker LH, Kadlubar FF, Lucier GW. Genotype/phenotype discordance for human arylamine N-acetyltransferase (NAT2) reveals a new slow-acetylator allele common in African-Americans. Carcinogenesis. 1993;14:1689-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 192] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Lee EJ, Zhao B, Seow-Choen F. Relationship between polymorphism of N-acetyltransferase gene and susceptibility to colorectal carcinoma in a Chinese population. Pharmacogenetics. 1998;8:513-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Fretland AJ, Leff MA, Doll MA, Hein DW. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 102] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol Rev. 1985;37:25-79. [PubMed] |
| 10. | Evans DA. N-acetyltransferase. Pharmacol Ther. 1989;42:157-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 183] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Zhao B, Seow A, Lee EJ, Lee HP. Correlation between acetylation phenotype and genotype in Chinese women. Eur J Clin Pharmacol. 2000;56:689-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Barrett JH, Smith G, Waxman R, Gooderham N, Lightfoot T, Garner RC, Augustsson K, Wolf CR, Bishop DT, Forman D. Investigation of interaction between N-acetyltransferase 2 and heterocyclic amines as potential risk factors for colorectal cancer. Carcinogenesis. 2003;24:275-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Schnakenberg E, Ehlers C, Feyerabend W, Werdin R, Hübotter R, Dreikorn K, Schloot W. Genotyping of the polymorphic N-acetyltransferase (NAT2) and loss of heterozygosity in bladder cancer patients. Clin Genet. 1998;53:396-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |