Published online Jun 21, 2005. doi: 10.3748/wjg.v11.i23.3504
Revised: May 26, 2004
Accepted: June 24, 2004
Published online: June 21, 2005
AIM: To evaluate the inhibitory effects of DNAzymes on the expressions of hepatitis B virus (HBV) s (HBsAg) and e (HBeAg) in 2.2.15 cells, and to explore the potential therapeutic effects of DNAzymes on replication of HBV genome.
METHODS: DNAzymes DrzBS and DrzBC specific to HBV (ayw subtype) s gene ORF A157UG and e gene ORF A1816UG, were designed and synthesized. Inhibitory effects of DrzBS or DrzBC on the expressions of HBV s and e genes as well as HBV DNA levels in culture supernatants were observed in 2.2.15 cells.
RESULTS: After being treated with DrzBS or DrzBC, the expression of HBV s or e genes in 2.2.15 cells was depressed dramatically. The maximum inhibition rate was 94.2% and 91.8% for DrzBS and DrzBC, respectively. The concentration for effective inhibition of both DrzBS and DrzBC was within 0.1-2.5 µmol/L, showing a dose-dependence. The efficiency of inhibiting HBsAg, HBeAg in 2.2.15 cells by DrzBS or DrzBC was higher than that of the same target genes by antisense oligonucleotides (ASON). The concentration for effective inhibition of DNAzymes was at least 10-fold lower compared with ASON controls. Neither inhibition on the replication of HBV DNA nor toxicity to 2.2.15 cells was observed.
CONCLUSION: DrzBS and DrzBC can block the expression of HBV s- and e-genes in 2.2.15 cells and provide a specific and effective anti-HBV gene therapeutic means.
- Citation: Wo JE, Wu XL, Zhou LF, Yao HP, Chen LW, Dennin RH. Effective inhibition of expression of hepatitis B virus genes by DNAzymes. World J Gastroenterol 2005; 11(23): 3504-3507
- URL: https://www.wjgnet.com/1007-9327/full/v11/i23/3504.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i23.3504
DNAzyme or deoxyribozyme is a specifically structured DNA sequence that possesses catalytic RNA-cleaving activity, recognizing its target RNA in a highly sequence-specific manner, and thereby blocks the expression of corresponding RNAs. mRNAs from oncogenes and viral genomes and transcripts are ideal targets for DNAzyme therapeutic agents. In 1997, Santoro and Joyce[1] proposed a universal DNAzyme structure termed DNAzyme 10-23, which consists of a catalytic domain in the center and recognition arms on both sides. Through its recognition arms, DNAzyme 10-23 combines specifically with target RNA sequences while the central catalytic domain sequentially cleaves the opposite site of the target RNA strands.
In this paper we report an observation of designed DNAzymes on the 2.2.15 cells, which is a part of a project aimed at DNAzyme inhibition of various viral gene expressions, such as hepatitis B virus (HBV), HCV, HIV and CMV, etc. Basically, we investigated in vitro cleavage effects of the designed DNAzymes on RNA substrates prepared by in vitro transcription in the presence of [a-32P]-ATP. The substrates were cleaved by the corresponding DNAzymes generating two labeled products of the correct size as judged by their electrophoretic mobility. Accordingly, we calculated the values of Km of the definite DNAzymes under different reaction conditions. Besides, we designed DNAzymes of different lengths of their arms targeting different sites. Details on the Km values and other topics which characterize the DNAzymes involved in this report will be summarized in an additional report. Here we present data of their effectiveness in 2.2.15 cell culture model.
2.2.15 cells were propagated in DMEM medium supplemented with 10% fetal bovine serum and 400 mg/L of G418.
Radio immunoassay (RIA) was used for the quantitative detection of HBsAg and HBeAg in cell culture medium with RIA kits (3V Diagnostic Tech Co., Shandong, China), according to the manufacturer’s instructions.
Fluorescence quantitative polymerase chain reaction (PCR) was used to determine the copies of HBV DNA in the cell culture medium with HBV nuclear acids determination kit (Piji Bioengineer Co., Ltd, Shenzhen, China) using the LightCycler PCR thermocycler (Roche Molecular Biochemics), according to the manufacturer’s instructions.
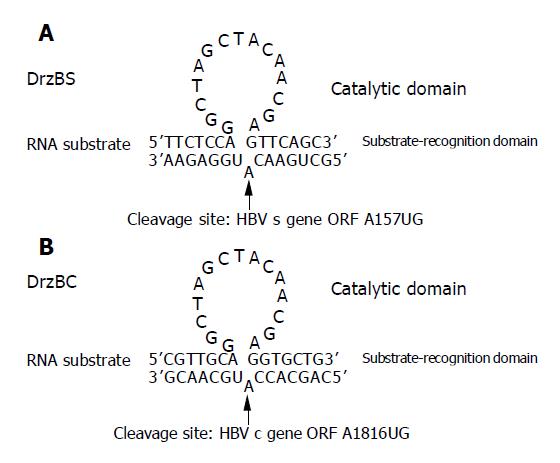
Nucleotide (nt) sequences of DNAzymes, antisense oligonucleotide (ASON) and nonsense oligonucleotide (NSON) controls targeted sequence sections within the HBV sequence region coding for HBs-antigen and the HBe-antigen respectively. Sequence data were shown in Table 1.
| DNAzyme | DrzBS | 5’TTCTCCAGGCTAGCTACAACGAGTTCAGC3’ |
| ASON | AsBS | 5’TTCTCCATGTTCAGC3’ |
| DNAzyme | DrzBC | 5’CGTTGCAGGCTAGCTACAACGAGGTGCTG3’ |
| ASON | AsBC | 5’CGTTGCATGGTGCTG3’ |
| NSON | Control | 5’ACGTCGTTGAAGCTAGCTGCACTGCTGGA3’ |
AGGCTAGCTACAACGAG was the catalytic domain sequence of DrzBS and DrzBC, while both its side sequences (underlined) were 6-mer recognizing arms, which specifically recognized the 6-nucleotide sequences just upstream and downstream of the starting AUG code of the HBV ayw subtype s gene or the c gene (GenBank accession no. X65257). Thus, the cleaving sites of the designed DrzBS and DrzBC targeted A157UG or A1816UG of the starting AUGs of the s or c gene ORFs (Figure 1). Two ASONs, AsBS and AsBC consisted of the same recognizing sequences as DrzBS, DrzBC and had the same starting AUGs in the middle. They were fully complementary to the sequences 150th-164th nt of the s gene and the 1809rd-1823rd nt of the c gene, functioning as controls for the recognizing arms of DrzBS and DrzBC. NSON was of the same length in nucleotides as DNAzymes designed randomly and showed no homology either to HBV genomes or to human genomic sequences. They were used as an oligonucleotide control for the 2.2.15 cells. All these DNAzymes, ASONs and NSON were synthesized by Shengong Bioengineer Co., Shanghai, China.
2.2.15 cells incubated in 24-well plates. DNAzymes were mixed with liposome (LipofectAMINE, Gibco BRL) 30 min before administration of cells, according to the manufacturer’s instructions. Culture supernatants collected at various time points following administration of liposome-coated DNAzymes were used for the determination of HBsAg and HBeAg concentrations. Three wells were used for each dose of DNAzymes and the concentrations of HBsAg or HBeAg were represented as averages from these three wells. The inhibitory effects induced by DNAzymes were evaluated according to the following equation.
Inhibition rate (IR) = |X of wells for DNAzyme admission-X of wells for control|/(X of wells for control-2.1) ×100%
where X represents the average S/P (RIA) value from three wells in HBsAg or HBeAg determination.
2.2.15 cells were observed for their growing features, and stained with 0.5% trypan blue to determine the number and ratio of stained living cells.
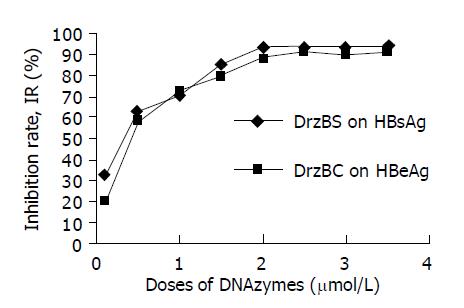
2.2.15 cells were propagated in plate wells for 24 h before administration of DNAzymes and controls. Forty-eight hours after administration of various concentrations of DrzBS and DrzBC, the culture medium was collected for HBsAg and HBeAg detection. Within a concentration range of 0.1-2.5 µmol/L, DrzBS increased IR of HBsAg expression from 32.4% to 94.2%, while DrzBC increased IR of HBeAg expression from 19.6% to 91.8%, showing a distinct dose-dependent effectiveness. With additional increases of DNAzyme admission doses, the IRs increased a little but platform-like effectiveness was observed (Figure 2). As controls for the ASONs, AsBS or AsBC exhibited dose-dependent inhibition effects within concentration ranges of 2.0-20 or 2.0-25 µmol/L for the expression of HBsAg or HBeAg respectively, while NSON as nonsense oligonucleotide control showed few inhibition effects within 2.0-25 µmol/L on the HBsAg and HBeAg expressions (all IRs <5%).
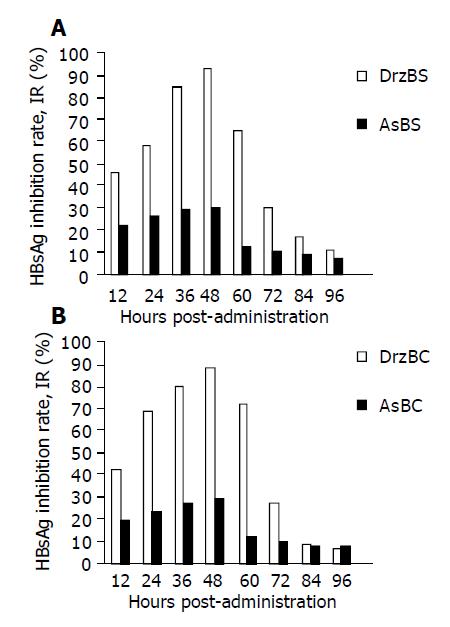
2.2.15 cells were primarily treated with DNAzymes at the concentration of 2.0 µmol/L. After 96 h, the culture medium in the wells was collected at 12-h intervals and supplemented with an equal amount of fresh medium without any DNAzymes. The time-efficiency characteristics of HBsAg and HBeAg suppression resulting from the DNAzyme administration are shown in Figure 3. Twelve hours after administration of DNAzyme, both DrzBS and DrzBC began to show their suppressing effect on the expression of HBsAg or HBeAg. The maximum suppression (IR = 94.2% for HBsAg and 91.8% for HBeAg) occurred after 48 h and decreased gradually thereafter possibly due to the decay of DNAzymes within cytoplasmic environment. After 72 h, the IR was 28.6% for HBsAg and 27.5% for HBeAg, and after 84 and 96 h. As controls, both AsBS and AsBC produced a lower IR of HBsAg or HBeAg expression at the same concentration of 2.0 µmol/L. Their IR also peaked after 48 h, but was just 30.2% for HBsAg and 29.3% for HBeAg, and was below 20% after 60 h (Figure 3).
Within the concentration of 0.1-2.5 µmol/L during a period of 96 h, no distinct change in HBV DNA copy number was observed (data not shown), indicating that HBV DNA excreted into the culture supernatants was not influenced by DNAzymes.
Within the concentration range and exposure periods mentioned above, the number of trypan blue-stained 2.2.15 cells remained stable (data not shown), and no obvious cytomorphological abnormalities were observed.
DNAzyme has been regarded as a breakthrough in revealing the traits of enzymes since the discovery of ribozyme[2]. Compared to ribozyme, DNAzyme has the following outstanding characteristics: 100000-fold more stable and resistant to degradation in the cytoplasmic environment[2], much simpler structure and thus easier to automatically synthesize on a large scale, much higher catalytic capacity 109/(mol•min)[1] and more specific to cleaving sites and RNA sequence-dependent cuttings. DNAzyme 10-23 i able to cleave A•U sites of RNA sequences, which means that theoretically it is able to cleave the starting AUGs of ORFs of any kind of mRNAs, thus providing a universal means of regulating and controlling the expression of all cellular proteins. Application of DNAzymes could facilitate new strategies for inhibiting the replication of RNA viruses, as well as the treatment of infectious diseases caused by RNA viruses. It has been reported that DNAzyme 10-23 specifically and successfully cleaves cellular mRNA strands deduced from certain genes and blocks their expression[3]. In the field of virology, DNAzyme 10-23 has been shown to cut efficiently and site-specifically gag/pol, env, vpr, tat and nef transcriptions of the human immunodeficiency virus type-1, to block the expression of corresponding genes and to inhibit viral replication in human papilloma virus E6/E7 transcription[2,4,5] .
In this study we tested deoxyribozymes against HBV rather than human hepatitis RNA viruses such as hepatitis A, C or E viruses (HAV, HCV or HEV). When the designed DNAzymes whose targets were specific to the HBV s or c gene were administrated to the 2.2.15 cells, expression of HBsAg or HBeAg was inhibited in a dose-dependent manner at the concentration of 0.1-2.5 µmol/L. At the concentration of 2.0 µmol/L, the levels of HBsAg or HBeAg were decreased by more than 90% compared to the blank controls. Such a distinct inhibition remained effective (IR>25%) for at least 72 h after administration of a single dose. Considering the observed lack of effect on the levels of HBV DNA excreted by the cells, it seems that the target molecules of designed DNAzymes are HBV RNA intermediates rather than HBV genomic DNA. Our results showed that the HBV-specific DNAzyme 10-23 structures were able to efficiently block the expression of HBV e- and s-genes within cells, resulting in the suppression of corresponding antigen proteins, suggesting that DNAzymes can be developed into a powerfully specific and effective form of anti-HBV gene therapy.
The substrate-specific cleavage of DNAzyme 10-23 depends on the nucleotide sequence of its recognition arms, i.e., cleavage occurs when the recognition arms bind to complementary nucleotide sequences of the target RNA strands. In order to ascertain that the observed inhibition did not result from possible antisense-blocking of its recognition sequences, we designed ASON controls (AsBS and AsBC) that consisted of the same nucleotide sequences as the DNAzymes’ recognition sequences. The concentration (0.1-2.5 µmol/L) for effective inhibition induced by DNAzymes was almost 10-fold lower than that of the assay with ASON AsBS (2.0-20 µmol/L) and ASON AsBC (2.0-25 µmol/L). After 48 h the IR of both DNAzymes at the concentration of 2.0 µmol/L on HBV antigens was more than 90%, while that of the ASONs was only about 30%. These results indicate that the designed DNAzymes can block the expression of HBV genes mainly through their site-specific cleaving effects, rather than through their recognition arms.
Although all the 2.2.15 cells were used in these experiments, the full length HBV genome was integrated. There are different ways of explaining this situation with ongoing HBV-DNA production. The cleaving is not complete, thus allowing some full length RNA transcripts to be used by the RT. The main targets for DNAzymes applied represent processed short mRNA molecules used for translation of respective ‘antigens’ only[6]. This situation of ongoing HBV-DNA production has been described when antisense DNA is used to inhibit viral antigen expression[7,8]. Further studies are required to reveal the background of this particular phenomenon.
| 1. | Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262-4266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1073] [Cited by in RCA: 1143] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 2. | Breaker RR. Tech.Sight. Molecular biology. Making catalytic DNAs. Science. 2000;290:2095-2096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 3. | Khachigian LM. Catalytic DNAs as potential therapeutic agents and sequence-specific molecular tools to dissect biological function. J Clin Invest. 2000;106:1189-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Cairns MJ, Hopkins TM, Witherington C, Wang L, Sun LQ. Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol. 1999;17:480-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 109] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 5. | Unwalla H, Banerjea AC. Inhibition of HIV-1 gene expression by novel macrophage-tropic DNA enzymes targeted to cleave HIV-1 TAT/Rev RNA. Biochem J. 2001;357:147-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Morrissey DV, Lee PA, Johnson DA, Overly SL, McSwiggen JA, Beigelman L, Mokler VR, Maloney L, Vargeese C, Bowman K. Characterization of nuclease-resistant ribozymes directed against hepatitis B virus RNA. J Viral Hepat. 2002;9:411-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 940] [Article Influence: 24.7] [Reference Citation Analysis (0)] |