Published online Jun 7, 2005. doi: 10.3748/wjg.v11.i21.3212
Revised: June 8, 2004
Accepted: June 24, 2004
Published online: June 7, 2005
AIM: To examine the aberrant expression of fragile histidine triad (FHIT) gene and protein in gastric cancer, and to evaluate the role of FHIT gene and the relationship between FHIT gene and EBV infection in gastric carcinogenesis.
METHODS: FHIT transcripts were detected by nested RT-PCR in 30 cases of gastric cancer and their products were sequenced. FHIT protein was detected by Western blot. EBV infection was detected by PCR method in 50 cases of gastric cancer.
RESULTS: The wild type transcripts were detected in all 30 matched normal tissues of gastric cancer. Aberrant transcripts were found in 11/30 (36.7%) gastric cancerous tissues. Sequencing analysis of the aberrant fragments found an RT-PCR product missing exons 5-7 in one case of gastric cancer, and another product missing exons 4-7. Four of ten (40.0%) cases of primary gastric cancer showed absent or decreased expression of FHIT protein as compared with their matched normal tissues. EBV was detected in 5/50 (10%) gastric cancers, among which 4/5 (80%) had aberrant transcripts of FHIT gene.
CONCLUSION: Loss of FHIT gene or FHIT protein plays an important role in carcinogenesis, development and progression of gastric cancer. EBV infection might influence carcinogenesis of gastric cancer by inducing the abnormality of FHIT gene.
- Citation: Xiao YP, Han CB, Mao XY, Li JY, Xu L, Ren CS, Xin Y. Relationship between abnormality of FHIT gene and EBV infection in gastric cancer. World J Gastroenterol 2005; 11(21): 3212-3216
- URL: https://www.wjgnet.com/1007-9327/full/v11/i21/3212.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i21.3212
Gastric carcinogenesis has been thought to be associated with its highly exposed risk factors such as Helicobacter pylori infection[1] and excessive nitrite salt intake[2-4]. In recent years, close attention has been paid to the carcinogenic factor of Epstein-Barr virus (EBV)[5-8]. It has been known that multiple oncogenes and tumor suppressor genes are involved in gastric carcinogenesis, however, no specific oncogenes and tumor suppressor genes have been identified. Fragile histidine triad (FHIT) gene, a tumor suppressor gene, is located at chromosome 3p14.2 and contains the most common aphidicolin-induced fragile site FRA3B[9]. FRA3B is the non-randomly fragmental or fissured site through chromosome’s spontaneous or induced actions, and also susceptible to chromosome loss and rearrangement because of the frequent fragmentation or fissure, which might be related to tumorigenesis[9]. Moreover, one of the EBV pathogenesis mechanisms might be related to the integration of EBV DNA into FRA3B and consequently decreasing FRA3B compliance[10]. Considering that FHIT contains the FRA3B region, EBV might make FHIT inactivated leading to carcinogenesis. In this paper, mRNA and protein expression of FHIT gene, EBV infection and their relationship in gastric carcinogenesis was determined.
Specimens, including 50 cases of primary gastric cancerous and matched distant normal gastric mucosal tissues, were surgically resected at the First Hospital of China Medical University without preoperative chemotherapy and radiotherapy. They were put immediately into liquid nitrogen, and reserved at -70 °C. All cases were classified respectively by Borrmann, WHO histological classification, Lauren classification, invasive depth and lymph node metastasis.
DNA and RNA extraction DNA was extracted from frozen gastric cancerous and normal mucosal tissues by standard phenol–chloroform method[6,8], dissolved and reserved in TE buffer[11]. RNA extraction was conducted according to instruction of TRIzol reagent kit (GIBCO). Briefly: frozen gastric tissues were ground into pieces, homogenized in l mL TRIzol reagent using a homogenizer for 30 s, and incubated for 5 min. and then 200 μL chloroform was added and spun by centrifugation at 3000 r/min for 15 min. The aqueous phase was transferred to another Eppendorf tube, 500 μL isoamyl alcohol was added and placed on ice for 1 h, and spun by centrifugation at 3000 r/min for 15 min, finally, the supernatant was discanded, RNA pellet was washed once with 750 mL/L ethanol, dissolved in 1 mL/L DEPC sterile water, and kept at -20 °C.
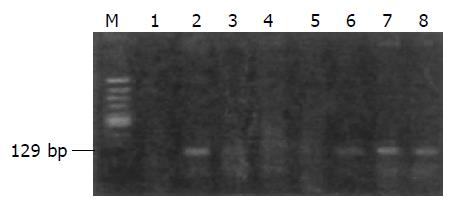
PCR and nested RT-PCR PCR and nested RT-PCR were used to amplify the EBV DNA and FHIT mRNA, respect-ively. Primer sequence for EBV was: (F)5’-CCAGACAGCA-GCCAATTGTC-3’; (R)5’-GGTAGAAGACCCCCTC-TTAC-3’. The PCR product was 129 bp in length, spanning the BamH1-w of EBV. PCR amplification was carried out in a final volume of 25 µL, containing 50 ng DNA, 0.5 µmol/L of each primer, 200 µmol/L of each dNTP, and 0.5 U Taq DNA polymerase (TaKaRa Ex TaqTM). The amplification condition: an initial incubation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min; and the final step of extension at 72 °C for 5 min. The cDNA synthesis of RT-PCR referred to the instruction of BcaBEST RNA PCR kit (TaKaRa). Nest-PCR primers were: (F2)5’-ATCCTGGAAGCTTTGAAGCTGA-3’; (R2)5’-TCACTGGTTGAAGAATACAGG-3’; (F1)5’-TCCGTA-*CTGCTATCTACATC-3’; (Rl)5’-CATGCTGATTCA-GTTCCTCTTGC-3’. First cycle of nest-PCR was the same as aforementioned except for PCR reaction volume 25 μL containing 50 ng cDNA, 0.5 mmol/L of each outer primer F2 and R2. The second cycle reaction system contained 5 μL of the first cycle product and 0.5 mmol/L of each inner primer F1 and R1, the others were similar.
Polyacrylamide gel electrophoresis (PAGE) EBV PCR products and nest-PCR products of FHIT gene in gastric cancerous and matched normal tissues were run on 1.5% agarose gel in 1×TBE buffer at a constant voltage of 60 V for 1 h.
Sequencing Abnormal nest-PCR products confirmed with agarose gel electrophoresis were cloned and sequenced by AoKe Biology Company (Beijing, China).
Protein extraction Tissues were sheared and homogenized in a homogenizer, added with suspending buffer containing 2 mmol/L EDTA, 10 mmol/L EGTA, 20 μmol/L Tris-HCl, pH 7.5, 56 g/L sucrose and 100 g/L PMSF, agitated with ultrasonic instrument, then spun by centrifugation at 4 °C 13000 g for 1 h. The supernatants were collected for Western blot.
Western blot Sixteen microliters of protein extracts were added into 4 μL 5×sampling buffer containing 130 mmol/L Tris-Cl, pH 8.0, 200 mL/L glycerol, 46 g/L SDS, 20 g/L DTT, pre-denatured at 96 °C for 10 min, run on 12% polyacrylamide gel (PAGE), then transferred to pyroxylin membrane at a voltage of 300 V for 2 h. The membrane was soaked in 1×TBS for approximately 10 min, and rabbit anti-human FHIT antibody (SANsc-8215, Gene Company Ltd) added at room temperature overnight, rinsed with 1×TBS buffer, goat anti-rabbit IgG antibody (Zhongshan Company, Beijing, China) added, shaken gently for 1 h, and rinsed with 1×TBS again. Finally Horseradish peroxidase (HRP) was added for 30 min and color development was done with DAB for 15-30 min, and membrane was rinsed with distilled water to stop reaction.
χ2-test was applied with SPSS software (version 10.0 for Windows). A P value of less than 0.05 was considered statistically significant.
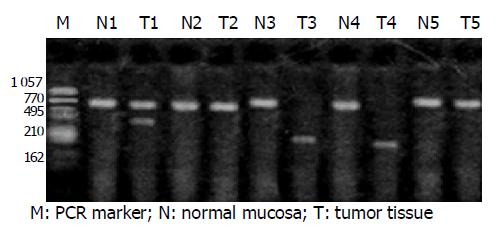
The normal transcripts (wild type, 707 bp) of FHIT gene were detected out in 30 cases of matched normal gastric mucosa. Abnormal transcripts were found in 11 of 30 cases (36.7%) of gastric cancers, among which five were shorter than 500 bp in length. Three cases of gastric cancers were simultaneously accompanied with normal transcripts, one with complete loss. Repeated experiments suggested that there existed FHIT gene loss in gastric cancer (Figure 1).
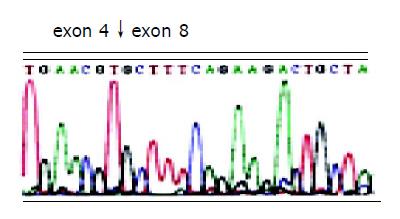
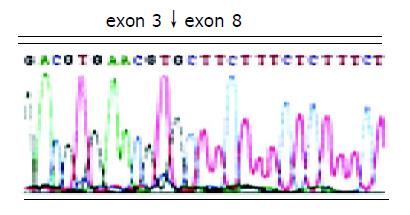
After two abnormal transcripts were purified via gel electrophoresis, the following sequencing results showed that one lost exons 5-7 of FHIT gene with a deletion of 297 bp and connection of exon 4 and exon 8; the other lost exons 4-7, with a deletion of 389 bp and connection of exon 3 and 8. In addition, all losses occurred in exon joints (Figures 2 and 3).
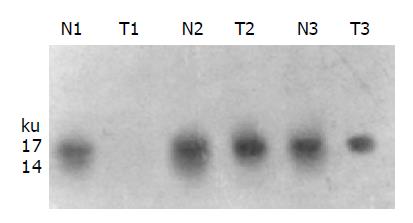
Western blot showed 4 of 10 cases (40.0%) of gastric cancers had loss or decreased expression of FHIT protein (Figure 4). EBV was detected in 5 of 50 (10%) cases of gastric cancers (Figure 5), among which 4 out of 5 (80.0%) has abnormal FHIT gene transcripts. The relationship between EBV infection and FHIT gene expression in gastric cancer is shown in Table 1.
| EBV | n | FHIT Transcripts | |
| Abnormal | Normal | ||
| + | 5 | 4 | 1 |
| - | 45 | 7 | 38 |
| Sum | 50 | 11 | 39 |
Exon 5 of FHIT gene is the first coding exon (initiation of translation), adjacent to fragile site FRA3B. DNA repair and rearrangement of FRA3B fissured site result in deletion of FHIT gene exon. Exon 8 containing highly conserved region, is a combing site of zinc ion and also the important target region of FHIT gene loss. The coding protein of FHIT gene, FHIT, a typical dinucleotide triphosphoric acid hydrolase (Ap3A), could hydrolyze adenosine diphosphate into ADP and AMP[12]. FHIT combines with mRNA cap structure, making mRNA de-cap, loss of function, increasing the probability of tumorigenesis[12]. FHIT combines with its substrate and exerts tumor suppression by its compound, which might be a signaling molecule and has more important function than its hydrolase[13].
Our research showed that FHIT gene in 36.7% gastric cancers presented truncated or deleted transcripts, about 450, 350, and 280 bp in length, suggesting there were different deletions of FHIT gene in gastric cancer. Sequencing results showed FHIT gene had deletion of exons 5-7 and 4-7. The deleted sites of FHIT transcripts were all splicing sites of FHIT gene exons, suggesting that deletion of FHIT gene usually resulted from its abnormal splice.
Baffa et al[14], reported that 53% gastric cancers had abnormal transcripts of FHIT gene, and 67% showed loss expression of FHIT protein. Huiping et al[15] , also reported that 87% gastric caners presented abnormal transcripts of FHIT gene. Ohta et al[16], discovered via nest RT-PCR that 53.6% gastric cancers and 37.5% large intestinal cancers had abnormal transcripts of FHIT gene, suggesting deletion of FHIT gene was common in gastric cancer. Abnormal transcripts of FHIT gene often involved one or more exons’ deletion, and mostly occurred in the two important functional exons 5 and 8, which were defined as types I and II, respectively by Ohta et al[16]. Whether types I or II, abnormal transcripts could not encode fully functional FHIT protein, so the two exons’ deletion might play a very important role in carcinomatous change of gastric mucosal epithelium. We summarize the abnormalities of FHIT gene transcripts as follows: (1) Exons 4-6 deletion; (2) exons 4-8 deletion; (3) single one exon deletion such as exon 5 or exon 8; (4) whole transcript’s deletion. The deletion of FHIT transcripts often involved important functional exons 5 and 8, further influencing the function of FHIT protein, resulting in a decrease or loss of FHIT protein activity, which affected catabolism of Ap3A and Ap4A. Accumulated intracellular Ap3A and Ap4A could repress cell apoptosis, and induce cell carcinomatous change[17-20].
Previous reports[21,22] showed that frequent deletions of FHIT transcripts were accompanied with decrease or loss of FHIT protein. Our researches showed that 40% gastric cancers displayed decreased or depleted FHIT protein (statistic analysis was not performed for fewer cases), and all of them had lymph node metastasis, suggesting that a decreased FHIT protein might be associated with lymph node metastasis and poor prognosis. Otterson et al[23], found that over-expression of FHIT protein could not suppress the cell clonal formation and cell proliferation. When transfer of normal FHIT gene into cancer cells without wide type FHIT per se, the product FHIT protein, however, could not suppress the cell growth. We presumed that the molecular mechanism of FHIT gene might be intrinsically different from the traditional tumor suppressor gene such as p53.
It has been found[5-7,24,25] that some gastric carcinomas are associated with the infection of EBV since Shibata et al[26], firstly reported their relationship. Most of the gastric carcinomas associated with EBV (EBVaGC) occurred at cardia or body of the stomach, or just appeared in the remnant gastric cancer, and showed the malignant features such as poor differentiation and infiltrated lymph nodes. We found five cases of gastric cancers infected with EBV and simultaneously all of them had lymph node metastasis, among which three cases were poorly differentiated late-stage gastric cancers, one moderately differentiated early-stage, and one poorly differentiated middle-stage gastric cancer. From the view of clinicopathological features, EBVaGC tended to be poorly differentiated, with lymph node metastasis, suggesting a poor prognosis of gastric cancer.
Divergent opinions have been reported concerning the molecular mechanism of EBVaGC. Some researchers[27-29] proposed that EBVaGC was associated with the abnormality of P53, Bcl-2, and TGF-β, and microsatellite instability. Chang et al[30], thought that there was no correlation between the infection of EBV and gastric carcinomas. At present, the mechanism of EBV in promoting gastric carcinogenesis has been unknown. Ohta et al[16], discovered that abnormalities of FHIT were frequently observed in some malignant tumors in certain areas, such as nasopharyngeal carcinomas in China and gastric cancers in Southeast Asia, and EBV might result in abnormal FHIT gene by its integration into the region of FRA3B. Four of the five gastric cancers with EBV infection in our study presented abnormal transcripts, indicating the close association between EBV and the abnormality of FHIT gene, and even the cell carcinomatous change. We also detected the loss of heterozygosity (LOH) and microsatellite instability (MSI) in the same five cases. LOH was observed at microsatellite sites D3S4103 (4/5), D3S1300 (3/5), D3S1481 (2/5) and D3S1234 (2/5), while MSI appeared at sites D3S1300 (1/5), D3S1481 (1/5) and D3S1234 (1/5). As D3S4103, D3S1300 and D3S1481 all located near exon 5 of FHIT gene, we presume that the integrating site of EBV might be just near exon 5.
In conclusion, we supply some new clues for further studies on the relations between abnormal FHIT and the malignant biological behaviors of gastric cancer cells. The studies of the relationship between EBV infection and abnormality of FHIT gene also provide a new scientific basis on pathogenesis of gastric cancer. By a certain mechanism such as integration, EBV might result in FHIT gene instability, down-regulate transcripts or truncated (deleted) FHIT proteins, which are likely to play an important role in gastric carcinogenesis, local infiltration and regional lymph node metastasis.
| 1. | Nardone G, Rocco A, Vaira D, Staibano S, Budillon A, Tatangelo F, Sciulli MG, Perna F, Salvatore G, Di Benedetto M. Expression of COX-2, mPGE-synthase1, MDR-1 (P-gp), and Bcl-xL: a molecular pathway of H pylori-related gastric carcinogenesis. J Pathol. 2004;202:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Mayne ST, Risch HA, Dubrow R, Chow WH, Gammon MD, Vaughan TL, Farrow DC, Schoenberg JB, Stanford JL, Ahsan H. Nutrient intake and risk of subtypes of esophageal and gastric cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:1055-1062. [PubMed] |
| 3. | Palli D. Epidemiology of gastric cancer: an evaluation of available evidence. J Gastroenterol. 2000;35 Suppl 12:84-89. [PubMed] |
| 4. | van Loon AJ, Botterweck AA, Goldbohm RA, Brants HA, van Klaveren JD, van den Brandt PA. Intake of nitrate and nitrite and the risk of gastric cancer: a prospective cohort study. Br J Cancer. 1998;78:129-135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 75] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Sudo M, Chong JM, Sakuma K, Ushiku T, Uozaki H, Nagai H, Funata N, Matsumoto Y, Fukayama M. Promoter hypermethylation of E-cadherin and its abnormal expression in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2004;109:194-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJ, Middeldorp JM, van den Brule AJ, Meijer CJ, Bloemena E. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol. 2004;22:664-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 216] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 7. | Oh ST, Seo JS, Moon UY, Kang KH, Shin DJ, Yoon SK, Kim WH, Park JG, Lee SK. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology. 2004;320:330-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Lee HS, Chang MS, Yang HK, Lee BL, Kim WH. Epstein-barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with epstein-barr virus-negative carcinoma. Clin Cancer Res. 2004;10:1698-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Mimori K, Inoue H, Shiraishi T, Matsuyama A, Mafune K, Tanaka Y, Mori M. Microsatellite instability is often observed in esophageal carcinoma patients with allelic loss in the FHIT/FRA3B locus. Oncology. 2003;64:275-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Lee HS, Chang MS, Yang HK, Lee BL, Kim WH. Epstein-barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with epstein-barr virus-negative carcinoma. Clin Cancer Res. 2004;10:1698-1705. |
| 11. | Han CB, Zhao YJ, Li F, He Q, Ma JM, Xin Y. Quantitation and detection of deletion in tumor mitochondrial DNA by microarray technique. Zhonghua ZhongLiu ZaZhi. 2004;26:10-13. [PubMed] |
| 12. | Fouts RL, Sandusky GE, Zhang S, Eckert GJ, Koch MO, Ulbright TM, Eble JN, Cheng L. Down-regulation of fragile histidine triad expression in prostate carcinoma. Cancer. 2003;97:1447-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Sükösd F, Kuroda N, Beothe T, Kaur AP, Kovacs G. Deletion of chromosome 3p14.2-p25 involving the VHL and FHIT genes in conventional renal cell carcinoma. Cancer Res. 2003;63:455-457. [PubMed] |
| 14. | Baffa R, Veronese ML, Santoro R, Mandes B, Palazzo JP, Rugge M, Santoro E, Croce CM, Huebner K. Loss of FHIT expression in gastric carcinoma. Cancer Res. 1998;58:4708-4714. [PubMed] |
| 15. | Huiping C, Kristjansdottir S, Bergthorsson JT, Jonasson JG, Magnusson J, Egilsson V, Ingvarsson S. High frequency of LOH, MSI and abnormal expression of FHIT in gastric cancer. Eur J Cancer. 2002;38:728-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3; 8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 736] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 17. | Chang KW, Kao SY, Tzeng RJ, Liu CJ, Cheng AJ, Yang SC, Wong YK, Lin SC. Multiple molecular alterations of FHIT in betel-associated oral carcinoma. J Pathol. 2002;196:300-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | An Q, Liu Y, Huang J. Comparison of tumor suppressor gene deletion between squamous cell carcinoma and adenocarcinoma lung cancer in Chinese. Zhonghua ZhongLiu ZaZhi. 2001;23:470-472. [PubMed] |
| 19. | Ko JY, Lee TC, Hsiao CF, Lin GL, Yen SH, Chen KY, Hsiung CA, Chen PJ, Hsu MM, Jou YS. Definition of three minimal deleted regions by comprehensive allelotyping and mutational screening of FHIT,p16(INK4A), and p19(ARF) genes in nasopharyngeal carcinoma. Cancer. 2002;94:1987-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Wistuba II, Ashfaq R, Maitra A, Alvarez H, Riquelme E, Gazdar AF. Fragile histidine triad gene abnormalities in the pathogenesis of gallbladder carcinoma. Am J Pathol. 2002;160:2073-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Andachi H, Yashima K, Koda M, Kawaguchi K, Kitamura A, Hosoda A, Kishimoto Y, Shiota G, Ito H, Makino M. Reduced Fhit expression is associated with mismatch repair deficiency in human advanced colorectal carcinoma. Br J Cancer. 2002;87:441-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Zhao P, Lu Y, Hu Y, Zhong M, Li Z, Li X. Loss of fragile histidine triad expression in colorectal carcinoma. Zhonghua BingLiXue ZaZhi. 2002;31:124-127. [PubMed] |
| 23. | Otterson GA, Xiao GH, Geradts J, Jin F, Chen WD, Niklinska W, Kaye FJ, Yeung RS. Protein expression and functional analysis of the FHIT gene in human tumor cells. J Natl Cancer Inst. 1998;90:426-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 224] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 25. | Kijima Y, Ishigami S, Hokita S, Koriyama C, Akiba S, Eizuru Y, Aikou T. The comparison of the prognosis between Epstein-Barr virus (EBV)-positive gastric carcinomas and EBV-negative ones. Cancer Lett. 2003;200:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Shibata DK. Epstein-Barr virus and gastric carcinoma. Jpn J Cancer Res. 1996;87:Inside front cover. [PubMed] |
| 27. | Sudo M, Chong JM, Sakuma K, Ushiku T, Uozaki H, Nagai H, Funata N, Matsumoto Y, Fukayama M. Promoter hypermethylation of E-cadherin and its abnormal expression in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2004;109:194-199. |
| 28. | zur Hausen A, Crusius JB, Murillo LS, Alizadeh BZ, Morré SA, Meijer CJ, van den Brule AJ, Peña AS. IL-1B promoter polymorphism and Epstein-Barr virus in Dutch patients with gastric carcinoma. Int J Cancer. 2003;107:866-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10:803-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 519] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 30. | Chang MS, Lee HS, Kim HS, Kim SH, Choi SI, Lee BL, Kim CW, Kim YI, Yang M, Kim WH. Epstein-Barr virus and microsatellite instability in gastric carcinogenesis. J Pathol. 2003;199:447-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |