Published online May 28, 2005. doi: 10.3748/wjg.v11.i20.3144
Revised: April 16, 2004
Accepted: May 9, 2004
Published online: May 28, 2005
AIM: To study the status of hMLH1 gene point mutations of gastric cancer kindreds and gastric cancer patients from northern China, and to find out gene mutation status in the population susceptible to gastric cancer.
METHODS: Blood samples of 120 members from five gastric cancer families, 56 sporadic gastric cancer patients and control individuals were collected. After DNA extraction, the mutations of exon 8 and exon 12 of hMLH1 gene were investigated by PCR-SSCP-CE, followed by DNA sequencing.
RESULTS: In the five kindreds, the mutation frequency was 25% (5/16) for the probands and 18% (19/104) for the non-cancerous members, which were significantly higher than the controls (P<0.01 χ2 = 7.71, P<0.01 χ2 = 8.65, respectively). In the sporadic gastric cancer, the mutation frequency was 7% (4/56), which was similar to that (5/100) in the healthy controls. The mutation point of exon 8 was at 219 codon of hMLH1 gene (A-G), resulting in a substitution of Ile-Val (ATC-GTC), whereas the mutation of exon 12 was at 384 codon of hMLH1 gene (T-A) resulting in a substitution of Asp-Val (GTT-GAT), which were the same as previously found in hereditary nonpolyposis colorectal carcinoma.
CONCLUSION: The members of gastric cancer families from northern China may have similar genetic background of hMLH1 gene mutation as those of hereditary nonpolyposis colorectal carcinoma.
- Citation: Li JH, Shi XZ, Lü S, Liu M, Cui WM, Liu LN, Jiang J, Xu GW. HMLH1 gene mutation in gastric cancer patients and their kindred. World J Gastroenterol 2005; 11(20): 3144-3146
- URL: https://www.wjgnet.com/1007-9327/full/v11/i20/3144.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i20.3144
Gastric cancer is one of the most common cancers and a leading cause of cancer death in China. Although the molecular biological mechanism involved in the gastric tumorigenesis remains unclear, clinical and epidemic studies have shown that gastric cancer occurrence has a tendency of familial accumulation. Several members can be attacked by gastric cancer just in one family. Moreover, the younger generations of these families are found to develop colorectal cancer at a high frequency in recent years because of the change of food habit. So we imagine that individuals susceptible to gastric cancer may have the similar genetic background as those of colorectal cancer. Hereditary nonpolyposis colorectal carcinoma (HNPCC) is a kind of colorectal cancer that has been well studied[1,2]. One of the reasons why HNPCC emerges is germline mutation in mismatch repair genes[3,4], most commonly in hMLH1 (human MutL homolog1) and hMSH2 (human MutS homolog2)[5]. Therefore, we selected the hot mutant sequences exon 8 and exon 12 of hMLH1 gene to detect gene mutation status in the population susceptible to gastric cancer.
Peripheral blood samples of 120 members in five gastric cancer families, 56 sporadic gastric cancer patients and control individuals were collected from the southern district of Liaoning Province, northeast of China. All blood samples were stored at -20 °C until used. Of the 120 familial samples, 16 were from gastric cancer patients (all were adenocarcinomas) aged 36-58 years, nine male and seven female and 104 were from non-cancerous members aged 9-70 years, 59 male and 45 female. All the five gastric cancer families met the following standards: three or more cases were affected by gastric cancer in two successive generations, two or more of them were first-degree relatives and at least one diagnosed to have had gastric cancer before the age of 50, whereas there was no first-degree relative affected by gastric cancer in those sporadic patients (all were adenocarcinomas) aged 42-64 years, 36 male and 20 female and healthy controls aged 18-60 years, 60 male and 40 female.
Genomic DNA was extracted from blood using the DNA extraction kit (Huashun Co., Ltd, Shanghai).
Primer sequences used were as follows: for exon 8, 5’-AM-ACAGACTTTGCTACCAGGACTTG-3’ (Forward) and 5’-FAM-TGTCTTATCCTCTGTGACAATGG-3’ (Reverse) and for exon 12, 5’-FAM-CTCAGCCATGAGAC-AATAAATCC-3’ (Forward) and 5’-FAM-GGTTCCCAAA-TAATGTGATGG-3’ (Reverse). Fluorescence-labeled primers and PCR amplification kits were obtained from TaKaRa Biotechnology (Dalian, China). The reaction mixture contained 10 mmol/L Tris-HCl (pH 9.0), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 200 µmol/L dNTPs, 1 mmol/L each primer, 100-200 ng of the extracted genomic DNA and 1.25 U Taq DNA polymerase in a total volume of 25 µL. PCR was performed with a Perkin-Elmer Model 2700 PCR system (Foster City, CA, USA) with the following polymerase chain reaction (PCR) program: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 40 s, annealing at 55 °C for 40 s, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 5 min.
Single-strand conformation polymorphism (SSCP) of DNA was analyzed by the method of capillary electrophoresis (CE). A P/ACE MDQ capillary electrophoresis instrument (Beckman, Palo Alto, CA, USA) with argon ion LIF detector (λex = 488 nm, λem = 520 nm) was employed. The capillary was coated with linear polyacrylamide with an inner diameter of 75 µm and an effective length of 30 cm. Prior to electrophoresis, the PCR products were diluted 20-folds with water, and heated at 95 °C for 10 min, then immediately put into ice water and kept for 5 min. The samples were injected at reverse polarity of 5 kV for 5 s, and separation was carried out under constant voltage. Single-strand DNA fragments were detected by LIF detector and the data were collected and analyzed by MDQ software.
The samples suspected to be mutant by CE-SSCP were sequenced. The PCR for sequencing was performed using nonfluorescent forward primer and the Big Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, ABI Prism) under the following conditions: initial denaturation at 96 °C for 2 min, 25 cycles at 96 °C for 10 s, annealing at 50 °C for 5 s, extension at 60 °C for 2 min, and a final elongation step at 60 °C for 7 min, in a 2700 thermal cycler (Perkin-Elmer). The products were purified by an ethanol/NaAc method, and then CE-sequencing was conducted using ABI Prism 310 (Perkin-Elmer, ABI Prism).
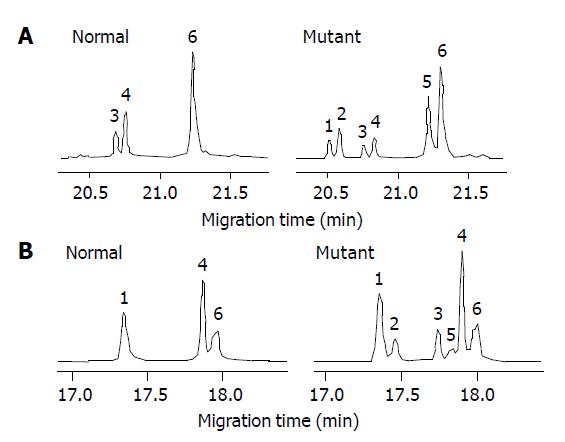
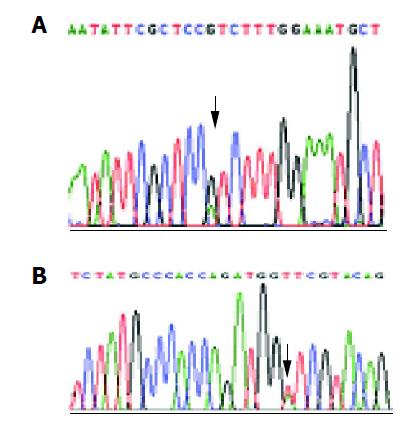
Of the 16 probands in the five kindreds, two cases were found mutant at exon 8, two cases were found mutant at exon 12 and the total mutation frequency was 25% (5/16). Meanwhile 12 cases were found mutant at exon 8, seven cases were found mutant at exon 12 and the total mutation frequency was 18% (19/104) in the familial noncancerous members. In the 56 cases of sporadic gastric cancer, two cases were found mutant at exon 8, two cases were found mutant at exon 12 and the total mutation frequency was 7% (4/56), which was similar to the healthy control individuals but being significantly lower than both the probands and familial members in the kindreds (Table 1). The mutation point of exon 8 was at 219 codon of hMLH1 gene (A-G), resulting in a substitution of Ile-Val (ATC-GTC) (Figures 1A and 2A), whereas the mutation of exon 12 was at 384 codon of hMLH1 gene (T-A), resulting in a substitution of Asp-Val (GTT-GAT) (Figures 1B and 2B), which was the same as previously found in HNPCC.
Mismatch repair (MMR) plays a central role in maintaining genomic stability by repairing DNA replication errors and inhibiting recombination between non-identical sequences[7,8]. Loss of mismatch repair causes destabilization of the genome and results in high mutation frequency[9]. HMLH1 gene is a prominent component in the human mismatch repair system and its dysfunction is involved in a number of patients with HNPCC[10,11]. It has been reported that germline mutations of MMR are identified in nearly 80% of the patients with HNPCC and almost 60% of the mutations are in hMLH1[5].
According to statistics, about 8-10% of gastric cancer cases are related to an inherited familial genetic factor[12]. Up to 3-fold increases in risk for gastric cancer among relatives of gastric cancer patients are consistent[13]. The gene background of gastric cancer susceptibility is not sufficiently known to us, especially in China, although the hereditary tendency is even higher.
We primarily detected exon 8 and exon 12 of the hMLH1 gene in 16 probands’ peripheral blood sample of five gastric cancer kindreds and found the same mutations that had previously been discovered in HNPCC. The mutation frequency was significantly higher than that of the control individuals and it was true of those familial members. It was proposed that the gene background is characterized by hMLH1 mutation in gastric cancer kindreds and that the presence of hMLH1 mutation is associated with an increased risk of developing gastric cancer and those carrying mutant hMLH1 gene are prone to develop gastric cancer even at an early age. What about sporadic gastric cancer? To answer this question, we extended the samples by analyzing 56 sporadic gastric cancer patients with their blood samples. To our surprise, the mutation frequency in the blood samples were similar to control individuals, which was significantly lower than both the probands and noncancerous members in the five kindreds’ blood samples. It seems that hMLH1 gene mutation is not a characteristic of sporadic gastric cancer.
From the above-mentioned, it can be concluded that there may be similar hMLH1 gene mutation as HNPCC in somatic cells in gastric cancer kindreds, but not in those in sporadic gastric cancer patients. The mutations of hMLH1 gene may be involved in tumorigenesis of hereditary gastric cancer in northern China.
| 1. | Park JG, Vasen HF, Park KJ, Peltomaki P, Ponz de Leon M, Rodriguez-Bigas MA, Lubinski J, Beck NE, Bisgaard ML, Miyaki M. Suspected hereditary nonpolyposis colorectal cancer: International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) criteria and results of genetic diagnosis. Dis Colon Rectum. 1999;42:710-715; discussion 715-716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Lynch HT, Watson P, Shaw TG, Lynch JF, Harty AE, Franklin BA, Kapler CR, Tinley ST, Liu B, Lerman C. Clinical impact of molecular genetic diagnosis, genetic counseling, and management of hereditary cancer. Part II: Hereditary nonpolyposis colorectal carcinoma as a model. Cancer. 1999;86:2457-2463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 3. | Dieumegard B, Grandjouan S, Sabourin JC, Le Bihan ML, Lefrère I, Bellefqih JP, Rougier P, Lasser P, Bénard J, Couturier D. Extensive molecular screening for hereditary non-polyposis colorectal cancer. Br J Cancer. 2000;82:871-880. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 61] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet. 1999;105:79-85. [PubMed] [Cited in This Article: ] |
| 5. | Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 518] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 6. | Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 364] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 7. | Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 153] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 8. | Duval A, Hamelin R. Genetic instability in human mismatch repair deficient cancers. Ann Genet. 2002;45:71-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [PubMed] [Cited in This Article: ] |
| 10. | Han HJ, Maruyama M, Baba S, Park JG, Nakamura Y. Genomic structure of human mismatch repair gene, hMLH1, and its mutation analysis in patients with hereditary non-polyposis colorectal cancer (HNPCC). Hum Mol Genet. 1995;4:237-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 113] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Hutter P, Couturier A, Membrez V, Joris F, Sappino AP, Chappuis PO. Excess of hMLH1 germline mutations in Swiss families with hereditary non-polyposis colorectal cancer. Int J Cancer. 1998;78:680-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | La Vecchia C, Negri E, Franceschi S, Gentile A. Family history and the risk of stomach and colorectal cancer. Cancer. 1992;70:50-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 13. | Zanghieri G, Di Gregorio C, Sacchetti C, Fante R, Sassatelli R, Cannizzo G, Carriero A, Ponz de Leon M. Familial occurrence of gastric cancer in the 2-year experience of a population-based registry. Cancer. 1990;66:2047-2051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |