Published online May 28, 2005. doi: 10.3748/wjg.v11.i20.3034
Revised: June 20, 2004
Accepted: July 22, 2004
Published online: May 28, 2005
AIM: To determine the features of microsatellite alterations and their association with clinicopathological characteristics of hepatocellular carcinoma (HCC).
METHODS: Loss of heterozygosity (LOH) and microsatellite instability (MSI) of 55 microsatellite loci were detected with PCR-based microsatellite polymorphism analyses in tumors and corresponding noncancerous liver tissues of 56 surgically resected HCCs using the MegaBACE 500 automatic DNA analysis system.
RESULTS: LOH was found in 44 of 56 HCCs (78.6%) at one or several loci. Frequencies of LOH on 1p, 4q, 8p, 16q, and 17p were 69.6% (39/56), 71.4% (40/56), 66.1% (37/56), 66.1% (37/56), and 64.3% (36/56), respectively. MSI was found in 18 of 56 HCCs (32.1%) at one or several loci. Ten of fifty-six (17.9%) HCCs had MSI-H. Serum HBV infection, alpha-fetoprotein concentration, tumor size, cirrhosis, histological grade, tumor capsule, as well as tumor intrahepatic metastasis, might be correlated with LOH on certain chromosome regions.
CONCLUSION: Frequent microsatellite alterations exist in HCC. LOH, which represents a tumor suppressor gene pathway, plays a more important role in hepatocarcin-ogenesis. MSI, which represents a mismatch repair gene pathway, is a rare event during liver carcinogenesis. Furthermore, LOH on certain chromosome regions may be correlated with clinicopathological characteristics in HCC.
- Citation: Zhang SH, Cong WM, Xian ZH, Wu MC. Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol 2005; 11(20): 3034-3039
- URL: https://www.wjgnet.com/1007-9327/full/v11/i20/3034.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i20.3034
Hepatocellular carcinoma (HCC) is one of the most frequent human cancers worldwide and has become the second cancer killer in China, since the 1990s[1]. Epidemiological studies in high-risk populations have identified chronic hepatitis B or C virus infections as well as dietary exposure to aflatoxin B1 as major factors in the etiology of this disease[2]. The genesis of human cancers is a multistep process reflecting cumulative genetic alterations that include activation of oncogenes or inactivation of tumor suppressor genes[3,4]. It has been proposed that genetic instability or genomic instability in human cancers can be divided into two types: chromosomal instability or loss of heterozygosity (LOH), which can result from errors in chromosome partitioning, and microsatellite instability (MSI), which is usually equated with DNA polymerase errors. Both LOH and MSI are considered to be phenotypes of genomic instability[5,6]. LOH is frequently observed on chromosomes 1p, 4q, 5q, 8p, 8q, 9p, 10q, 11p, 13q, 14q, 16q, 17p, and 22q in HCC, which suggests that tumor suppressor genes may take part in hepatocarcinogenesis[7-9]. MSI and mutations of mismatch repair gene have been reported in chronic hepatitis, cirrhosis and HCC in recent studies[10-14]. In the present study, we examined the genetic instability in 56 HCCs with 55 high-polymorphic microsatellite markers of chromosomes 1, 4, 8, 16, and 17, and analyzed the association of microsatellite alterations and the clinicopathological characteristics of HCC, in order to further understand the molecular mechanisms of hepatocarcinogenesis.
Fifty-six liver cancer specimens from surgically resected tissues were obtained from Eastern Hepatobiliary Surgery Hospital, Second Military Medical University. All patients have not received any prior therapy. All specimens were confirmed with histopathological examination. This study included 43 men and 13 women. The age ranged from 29 to 78 years. Thirty-five of fifty-six patients had serum alpha-fetoprotein (AFP) ≥20 µg/L. There were 39 cases positive for HBsAg. Twelve cases had small HCCs (≤3 cm) and 44 had advanced HCCs (>3 cm). Histopathological diagnosis was made according to the WHO histological classification of tumors of the liver and intrahepatic bile ducts (2000). Twenty HCCs were well differentiated, 28 were moderately differentiated and 8 were poorly differentiated. Of the 56 patients, 45 had evidence of intrahepatic metastasis (portal vein invasion and/or intrahepatic dissemination). Forty-seven HCCs were detected accompanying liver cirrhosis.
Fresh samples were obtained, immediately frozen in liquid nitrogen and stored at -80 °C until analysis. A microdissection technique with a cryostat was used to separate tumor cells from corresponding noncancerous liver tissues. Genomic DNA was extracted from carcinoma tissue and corresponding noncancerous liver tissues, using the standard phenol/chloroform method[15]. Concentration of DNA was determined with both spectrophotometric and fluorometric methods.
Fifty-five microsatellite loci used in this study with chromosomal positions are listed in Table 1. The chromosomal positions and sequences of each microsatellite marker could be referenced to the genome database (http://gdbwww.gdb.org/) and the co-operative human linkage center (http://www.chlc.org/). Each primer pair was fluorescent dye labeled. The PCR mixture contained more than 10 ng of genomic DNA, 200 µmol/L of each dNTP, 1.5 mmol/L MgCl2, 0.5 unit of AmpliTaq Gold DNA polymerase (PE Applied Biosystems, Foster City, CA, USA), 0.5 µmol/L of each primer, and 10×AmpliTaq Gold PCR buffer in a final volume of 10 µL. After denaturation at 94 °C for 12 min, DNA amplification was performed for 15 cycles at 94 °C for 30 s, at 63 °C for 60 s (decreased 0.5 °C of each cycle), and at 72 °C for 90 s; then for 25 cycles at 94 °C for 30 s, at 56 °C for 60 s, and at 72 °C for 90 s; with a final extension at 72 °C for 10 min. PCRs were run in a Biometra thermocycler (Biometra, Germany).
| Locus | Location | Allelic size(bp) | Heterozygosity (%) | Informative cases | LOH n(%) | MSI n(%) |
| D1S243 | 1p36.3 | 142–170 | 0.86 | 35 | 16(45.7) | 3(8.57) |
| D1S468 | 1p36.3 | 173–191 | 0.75 | 35 | 13(37.1) | 3(8.57) |
| D1S2893 | 1p36.2 | 201–223 | 0.47 | 44 | 18(40.9) | 2(4.55) |
| D1S2694 | 1p36.2 | 241–255 | 0.65 | 38 | 16(42.1) | 1(2.63) |
| D1S450 | 1p36.2 | 243–267 | 0.81 | 38 | 17(44.7) | 1(2.63) |
| D1S434 | 1p36.2 | 240–252 | 0.61 | 42 | 15(35.7) | 0 |
| RIZ | 1p36.2 | – | – | 30 | 23(76.7) | 0 |
| D1S507 | 1p36.1 | 183–203 | 0.78 | 45 | 24(53.3) | 2(4.44) |
| D1S199 | 1p36.1 | 94–116 | 0.84 | 39 | 14(35.9) | 1(2.56) |
| D1S234 | 1p36.1 | 268–294 | 0.5 | 37 | 14(37.8) | 3(8.11) |
| D4S1538 | 4q22 | 149–161 | 0.69 | 41 | 17(41.5) | 0 |
| D4S1534 | 4q22 | 146–158 | 0.77 | 41 | 22(53.7) | 0 |
| D4S406 | 4q26 | 234–258 | 0.87 | 42 | 22(52.4) | 1(2.38) |
| D4S1625 | 4q27-31 | 182–210 | 0.74 | 38 | 13(34.2) | 1(2.63) |
| D4S1652 | 4q31.1 | 115–125 | 0.75 | 38 | 16 (42.1) | 0 |
| D4S1615 | 4q34-qter | 138–150 | 0.71 | 39 | 19(48.7) | 0 |
| D4S2361 | 4q | 149–164 | 0.7 | 42 | 15(35.7) | 1(2.38) |
| D4S1554 | 4q35 | 184–208 | 0.86 | 37 | 14(37.8) | 0 |
| D4S426 | 4q35 | 177–191 | 0.78 | 41 | 25(61) | 0 |
| D4S2921 | 4q35 | 141–163 | 0.56 | 44 | 23(52.3) | 2(4.55) |
| D8S264 | 8p23 | 121–145 | 0.83 | 43 | 19(44.2) | 1(2.32) |
| D8S277 | 8p23 | 148–180 | 0.73 | 38 | 18(47.4) | 1(2.63) |
| D8S1706 | 8p23 | 257–281 | 0.88 | 41 | 20(48.9) | 1(2.43) |
| D8S1721 | 8p23 | 170–212 | 0.71 | 40 | 21(52.5) | 0 |
| D8S520 | 8p23 | 179–199 | 0.77 | 40 | 20(50.0) | 2(5.00) |
| D8S549 | 8p22 | 166–172 | 0.61 | 44 | 20(45.5) | 2(4.55) |
| D8S261 | 8p22 | 128–144 | 0.79 | 43 | 23(53.5) | 1(2.33) |
| D8S298 | 8p22 | 155–167 | 0.68 | 44 | 23(52.3) | 1(2.27) |
| D8S1733 | 8p21 | 253–257 | 0.64 | 38 | 17(44.7) | 1(2.63) |
| D8S1771 | 8p21 | 218–240 | 0.77 | 40 | 21(52.5) | 1(2.50) |
| D16S408 | 16q13 | – | – | 41 | 17(41.5) | 1(2.22) |
| D16S512 | 16q22.1 | 201–211 | 0.77 | 45 | 20(44.4) | 6(13.3) |
| D16S515 | 16q22.1 | 222–244 | 0.81 | 36 | 12(33.3) | 2(5.56) |
| D16S507 | 16q24.1 | 175–195 | 0.78 | 44 | 20(45.5) | 4(9.09) |
| D16S534 | 16q24.2 | 296–364 | 0.87 | 42 | 21(50.0) | 5(11.9) |
| D16S3091 | 16q24.2 | 115–129 | 0.72 | 42 | 22(59.5) | 2(4.76) |
| D16S422 | 16q24.2 | 188–212 | 0.79 | 42 | 23(54.8) | 2(4.76) |
| D16S520 | 16q24.3 | 181–197 | 0.84 | 44 | 19(43.2) | 2(4.55) |
| D16S413 | 16q24.2 | 208–246 | 0.68 | 36 | 16(44.4) | 2(5.56) |
| D16S498 | 16q24.3 | 131–149 | 0.84 | 46 | 23(50.0) | 1(2.17) |
| D17S849 | 17p13.3 | 251–261 | 0.69 | 39 | 21(53.9) | 1(2.56) |
| D17S926 | 17p13.3 | 243–259 | 0.81 | 36 | 21(58.3) | 1(2.78) |
| D17S831 | 17p13.3 | 224–246 | 0.81 | 37 | 25(67.6) | 3(8.11) |
| D17S938 | 17p13.1-p13.3 | 164–182 | 0.76 | 42 | 21(50.0) | 0 |
| D17S786 | 17p13.1 | – | – | 39 | 15(38.5) | 1(2.56) |
| D17S520 | 17p13.1 | 135–157 | 0.77 | 40 | 15(37.5) | 2(5.00) |
| D17S799 | 17p13-p12 | – | 0.8 | 42 | 21(50.0) | 2(4.76) |
| D17S921 | 17p12 | 186–200 | 0.7 | 39 | 15(38.5) | 0 |
| D17S261 | 17p11.2 | 169–185 | 0.72 | 41 | 15(36.6) | 1(2.44) |
| TP53 | 17p11.2 | 157–171 | 0.54 | 7 | 4(57.1) | 0 |
| D2S123 | hMSH-2 | 197–227 | 36 | 3 | 7(19.4) | |
| BAT-26 | 116 | 37 | 2 | 9(24.3) | ||
| D5S346 | APC | 96–122 | 41 | 1 | 8(19.5) | |
| D13S170 | Rb | 220–240 | 32 | 4 | 7(21.9) | |
| D17S250 | 151–169 | 33 | 3 | 5(15.2) |
PCR products were purified by 70% alcohol, twice after amplification. The reaction conditions sometimes yielded an unsatisfactory amplification for certain markers and were modified, and if necessary, these particular markers were run singly. The injection mixture was prepared for each capillary buffer by adding the following to each well of the injection palate: 2 μL purified sample, 0.25 μL ET400-R size standard, and 2.75 μL loading solution (Amersham Pharmacia Biotech). The injection mixture was centrifuged and heat-denatured for 2 min at 94 °C, then immediately cooled and placed on ice. Samples were electrophoresed on the MegaBACE-500 capillary array electrophoresis sequencer, and the fluorescent signals from different sized alleles were recorded and analyzed using Genetic Profiler version 2.1 software.
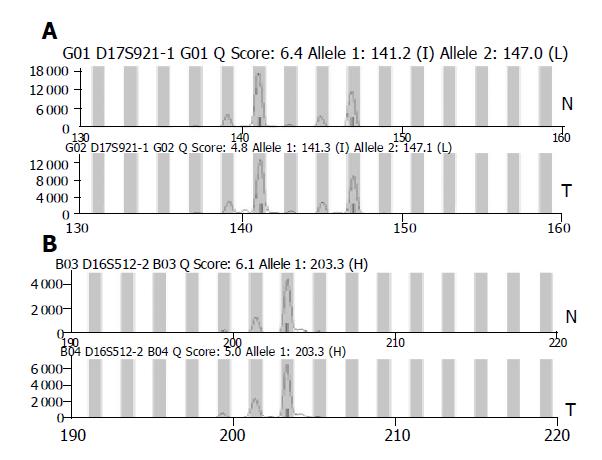
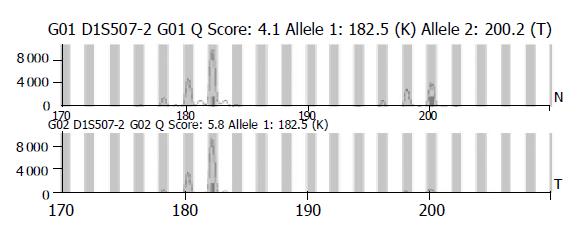
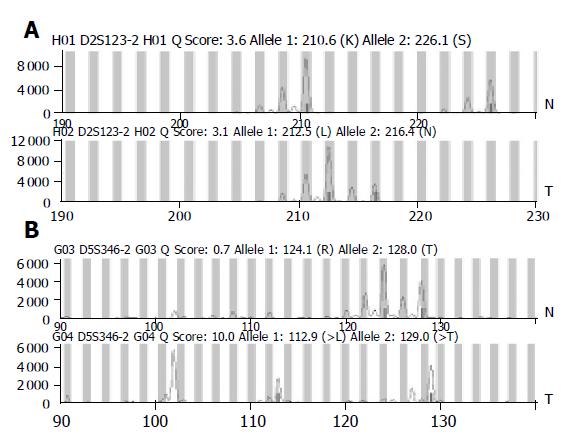
The existence of LOH or MSI was determined by comparing the area between cancerous tissues and corresponding noncancerous liver tissues. When two amplified bands per locus were detected in noncancerous liver tissue specimens, the case was defined as informative for analysis (Figure 1A). It was abandoned when only one amplified band per locus was detected in noncancerous liver tissue specimens (Figure 1B). For a given informative marker, LOH was identified when one of two bands was absent between the tumor DNA and normal DNA (Figure 2). MSI was assessed using the five markers recommended by the NCI workshop. MSI high (MSI-H) HCCs were defined as those having two unstable markers of the five markers (D2S123, BAT-26, D5S346, D13S170, and D17S250). MSI was defined as a band shift in one of the two alleles or the presence of novel bands in the two alleles or in the tumor DNA (Figure 3)[16]. Analysis of LOH and MSI was performed at least in duplicate.
The differences between the frequency of LOH and clinicopathological features were statistically analyzed using χ2 test. P<0.05 was considered statistically significant.
LOH was found in 44 of 56 HCCs (78.6%) at one or several loci. The frequency of LOH in HCC at 55 loci is shown in Table 1. Frequencies of LOH on 1p, 4q, 8p, 16q, and 17p were 69.6% (39/56), 71.4% (40/56), 66.1% (37/56), 66.1% (37/56), and 64.3% (36/56), respectively. The loci with the highest frequency of LOH were RIZ (76.7%) on 1p, D4S426 (61%) on 4q, D8S261 (53.5%) on 8p, D16S3091 (59.5%) on 16q and D17S831 (67.6%) on 17p. We performed MSI analysis using the five markers recommended by the NCI workshop. MSI was found in 18 of 56 HCCs (32.1%) at one or several loci. The frequency of MSI was as follows: 10 of 56 (17.9%) HCCs had MSI-H, 8 of 56 (14.3%) had MSI-L. The top two loci were BAT-26 (9/37, 24.3%) and D13S170 (7/32, 21.9%). All seven cases were positive serum HBsAg, of them five were positive for serum HBsAg. There was no significant difference in viral status between the cases with and without MSI. MSI-L was also found in some loci except these five loci.
To determine whether LOH was associated with clinicopath-ological features and to reveal its biological role in HCC development and/or progression, we compared LOH with clinicopathological findings including HBV infection, serum AFP concentration, tumor size, cirrhosis, histological grade, tumor capsule, and tumor intrahepatic metastasis. However, no statistically significant association was found between total LOH and clinicopathological findings. Correlation between clinicopathological features and LOH in HCC is shown in Table 2.
| Parameter | Locus | LOH/informative cases | P | |
| HBsAg | Positive | Negative | ||
| D1S2893 | 17/31 | 1/13 | 0.004 | |
| D1S507 | 13/21 | 1/16 | 0.001 | |
| D4S406 | 20/26 | 2/16 | 0.001 | |
| D8S277 | 16/22 | 2/16 | 0.001 | |
| D8S261 | 10/27 | 13/16 | 0.005 | |
| D8S298 | 11/29 | 12/15 | 0.008 | |
| D8S1733 | 7/24 | 10/14 | 0.011 | |
| D16S512 | 10/31 | 10/14 | 0.014 | |
| D16S515 | 5/27 | 7/9 | 0.001 | |
| D17S831 | 19/21 | 6/16 | 0.001 | |
| D17S938 | 19/25 | 2/14 | 0.001 | |
| D17S926 | 13/27 | 8/9 | 0.032 | |
| AFP (mg/L) | <20 | ≥20 | ||
| D1S507 | 3/17 | 21/29 | 0.001 | |
| D16S413 | 9/12 | 7/24 | 0.009 | |
| Liver cirrhosis | Present | Absent | ||
| D16S534 | 20/34 | 1/9 | 0.011 | |
| D16S3091 | 24/33 | 1/9 | 0.001 | |
| D16S422 | 22/34 | 1/8 | 0.008 | |
| D17S849 | 21/31 | 1/8 | 0.005 | |
| D17S831 | 24/39 | 1/8 | 0.011 | |
| D17S799 | 20/35 | 1/7 | 0.038 | |
| Tumor size | ≤3 cm | >3 cm | ||
| D8S298 | 2/9 | 21/35 | 0.043 | |
| D8S1771 | 0/9 | 21/31 | 0.001 | |
| D16S498 | 4/12 | 19/24 | 0.007 | |
| D17S926 | 3/11 | 18/25 | 0.012 | |
| Histological grade | Well | Moderate and poor | ||
| D4S426 | 2/11 | 23/30 | 0.002 | |
| D4S1615 | 2/12 | 14/26 | 0.04 | |
| D4S1652 | 2/11 | 17/28 | 0.031 | |
| D16S498 | 2/18 | 21/29 | 0.001 | |
| D17S926 | 4/13 | 17/23 | 0.012 | |
| Tumor capsule | Absent or not intact | Intact | ||
| D8S1721 | 20/28 | 1/12 | 0.001 | |
| Intrahepatic metastasis | Not observed | Observed | ||
| D1S468 | 8/10 | 5/25 | 0.001 | |
| D4S2921 | 2/21 | 21/23 | 0.023 | |
| D8S298 | 2/10 | 21/34 | 0.02 | |
| D8S1771 | 2/9 | 19/31 | 0.039 | |
| D17S926 | 1/11 | 20/25 | 0.001 | |
| D17S786 | 0/10 | 15/29 | 0.004 | |
Recently, genomic alterations in HCC have been studied by comparative genomic hybridization, and several chromosome changes have been reported. The gain of 1q, 8q, and 17q and the loss of 4q, 8p, 13q, 16q and 17p were commonly found in HCC. The chromosome regions with gain contained critical oncogenes, whereas those with loss contained tumor suppressor genes[17-19]. Frequent LOH on chromosomes 1p, 4q, 6p, 8p, 13q, 16q, and 17p by whole genome allelotyping suggested that putative tumor suppressor genes on these chromosome regions might play important roles in the development and progression of HCC[7-9].
Frequent deletions and translocations of the short arm of chromosome 1 (1p) have been found in HCCs by cytogenetic analysis, in situ hybridization, RFLP and microsatellite analysis, and mapped a commonly affected LOH region to 1p35-36[8,20]. In the present study, LOH was found in 39 of 56 HCCs (69.6%) on at least 1 locus, the top two loci were RIZ (76.7%) at 1p36.2 and D1S199 (53.3%) at 1p36.1. Early studies have demonstrated that high frequency of LOH on chromosome 4q in HCC was mapped to 4q11-q12, 4q12-23, 4q22-24 and 4q35[8,21]. Our data also showed that LOH was found in 71.4% HCCs on at least one locus, the top two loci were D4S426 (61%) at 4q35, and D4S1534 (53.7%) at 4q22. A high frequency of LOH on chromosome 8p has been reported in HCC and other cancers[22,23]. Nagai et al[8] reported that 42% of HCCs had LOH on 8p21-23 and argued that a tumor suppressor gene for HCC located in this region. Candidate tumor suppressor genes, such as PRLTS gene, DLC-1 gene, and EXTR1 gene located on 8p21-22, have been found to be mutated in a variety of tumors, including HCC, ovarian cancers, breast cancer and sporadic colorectal carcinoma[24-27]. Our study showed that LOH was found in 37 of 56 HCCs (66.1%) on at least 1 locus, the top three loci were D8S261 (53.5%) at 8p22, D8S1721 (52.5%) at 8p23 and D8S1771 (52.5%) at 8p21. The highest LOH regions were mapped to 16q12.1, 16q12.2 and 16q22-24[8,21,28]. Our data also showed that 71.4% HCCs harbored LOH on at least 1 locus, the alterations occurring in the top two loci were D16S3091 (59.5%) and D16S422 (54.8%) at 16q24.2. These regions contain many important genes, such as adenine phosphoribosyl transferase (APRT, 16q24.2) and N-acetylgalactosamine-6-sulfatase (GALNS, 16q24.2). The deletion of these genes in this region might interfere with cell growth or function[28]. LOH of 17p in 70% has been reported previously[29]. A missense mutation with replacement of arginine by serine at codon 249 has been reported previously in HCCs from Qidong, Shanghai and other geographical areas where aflatoxin and HBV are present[30]. Our data revealed a high level of LOH in D17S831 (67.6%) and D17S926 (58.3%) at 17p13.3. Zhao et al[31], also found a novel growth suppressor gene on chromosome 17p13.3 with a high frequency of mutation in human HCC. These results suggest that tumor suppressor genes at 1p36, 4q22, 4q35, 8p22-23, 16q24.2, and 17p13.3 may be involved in hepatocarcinogenesis.
HBV infection has been regarded as an important factor in the development of HCC[32]. However, the molecular mechanism is unclear. Wong et al[18], have assessed the genome-wide chromosomal analysis in 83 tumor samples from Hong Kong, Shanghai, Japan, and USA by comparative genomic hybridization. The most striking feature from the analysis was the high number of aberrations per sample in the HBV-related cases from Shanghai, which was significantly more than that in the other groups. Becker et al[33], have shown frequent loss of chromosome 8p in HBV-positive HCC from China. In this study, we found that the incidences of LOH on D1S2893, D1S507, D4S406, D8S277, D17S831, and D17S938 were significantly higher in patients with positive serum HBsAg than in those with negative HBsAg. These data imply that HBV infection might cause genetic changes within chromosomal regions where this is important for the development of some HCCs. Interestingly, LOH on D4S1538, D8S261, D8S298, D8S1733, D16S512, D16S515, and D17S926 occurred more frequently in HBsAg-negative patients than in HBsAg-positive patients. These results suggest that the tumor suppressor genes near these loci might be responsible for HBV-negative HCC.
Aggressive tumor phenotypes such as larger tumor size, poor cellular differentiation, absence of tumor encapsulation and intrahepatic metastasis were associated with LOH at several specific loci[34,35]. LOH was associated with an elevated serum AFP[21]. LOH on D8S298 at 8p22 was closely associated with venous permeation, tumor microsatellite formation, and larger tumor size[22]. LOH on locus D8S1721 at 8p23.1 was seen more frequently in nonencapsulated tumors and LOH on D8S1771 at 8p21.3 was associated with a larger tumor size and poorer cellular differentiation. LOH on D1S214 (lp36.3) and D1S2797 (1p34) was more frequently detected in tumors with intrahepatic metastasis than in those without[9]. In this study, large tumor size (>3 cm) tended to have a higher frequency of LOH on D8S298, D8S1771, D16S498, and D17S926. LOH on D4S426, D4S1615, D4S1652, D16S498, and D17S926 was more frequent in poorly or moderately differentiated HCC than in well-differentiated HCC. LOH frequencies of D8S1721 were significantly higher in patients without intact tumor capsule than in those with intact tumor capsule. LOH on D4S2921, D8S298, D8S1771, D17S926, and D17S786 were more frequently detected in tumors with intrahepatic metastasis than in those without. These results supported that 1p, 4q, 8p, 16q, and 17p deletions at specific loci were associated with tumor progression and aggressive behavior. Association of LOH at specific loci with a more aggressive tumor behavior suggests that loss/inactivation of putative tumor suppressor gene (s) located at these regions may confer a tumor growth advantage and contribute to the progression of HCC. This finding agrees with a previous report using CGH analysis, which suggested that the deletion was associated with tumor metastasis in HCC patients[35].
MSI with associated deficient DNA mismatch repair was first described in hereditary nonpolyposis colorectal cancer and has been implicated in the pathogenesis of a variety of gastrointestinal and other cancers[5]. It is noteworthy that there are some differences in MSI frequency of HCC reported in literatures. The MSI frequencies of HCC were 41-66.6% in Greece, USA and France[11,36]. However, Piao et al[37], and Yamamoto et al[38], reported the absence of MSI in Korean or Japanese HCCs, and considered that MSI played no role in the development or progression of HCC. Macdonald et al[39], found that the frequent LOH of MSI in HCC linked to hMSH2 and/or hMLH1 was only 19.6%, usually in association with MSI. Besides, another recent study on Chinese HCCs spanning 22 autosomes using 292 highly polymorphic markers showed that MSI was rarely seen[28]. In our study, MSI was found in 18 of 56 HCCs (32.1%) at one or several loci. Ten of fifty-six (17.9%) HCCs had MSI-H. Cases mostly were positive for serum HBsAg. Some studies showed that the incidence of MSI in cirrhotic liver tissues (27%) was almost three times higher than that in noncancerous liver tissues exhibiting findings compatible with chronic hepatitis (10%). In addition, the incidence of MSI in cirrhotic liver tissues did not differ from the incidence in HCCs[40]. The above-mentioned discrepancies may be partly due to the difference in selected cases, the sensitivity of methodology, and the type and number of polymorphic markers used in these studies. The other factors such as different geographic area and different criteria for MSI might contribute to the difference.
In conclusion, frequent microsatellite LOH on chromosomes 1, 4, 8, 16, and 17 existed in HCC. LOH, representing a tumor suppressor gene pathway, plays a more important role in hepatocarcinogenesis. MSI is less frequently found than LOH, suggesting a minor role of DNA mismatch repair deficiency in liver carcinogenesis.
| 1. | Tang ZY. Treatment of hepatocellular carcinoma. Digestion. 1998;59:556-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 671] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 3. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 4. | Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593-2604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 5. | Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci USA. 2003;100:776-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 485] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 6. | Kawai H, Suda T, Aoyagi Y, Isokawa O, Mita Y, Waguri N, Kuroiwa T, Igarashi M, Tsukada K, Mori S. Quantitative evaluation of genomic instability as a possible predictor for development of hepatocellular carcinoma: comparison of loss of heterozygosity and replication error. Hepatology. 2000;31:1246-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 245] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 8. | Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A. Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene. 1997;14:2927-2933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 204] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, Yu XJ, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J Hepatol. 2001;34:840-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Dore MP, Realdi G, Mura D, Onida A, Massarelli G, Dettori G, Graham DY, Sepulveda AR. Genomic instability in chronic viral hepatitis and hepatocellular carcinoma. Hum Pathol. 2001;32:698-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Karachristos A, Liloglou T, Field JK, Deligiorgi E, Kouskouni E, Spandidos DA. Microsatellite instability and p53 mutations in hepatocellular carcinoma. Mol Cell Biol Res Commun. 1999;2:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Maggioni M, Coggi G, Cassani B, Bianchi P, Romagnoli S, Mandelli A, Borzio M, Colombo P, Roncalli M. Molecular changes in hepatocellular dysplastic nodules on microdissected liver biopsies. Hepatology. 2000;32:942-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Roncalli M, Bianchi P, Grimaldi GC, Ricci D, Laghi L, Maggioni M, Opocher E, Borzio M, Coggi G. Fractional allelic loss in non-end-stage cirrhosis: correlations with hepatocellular carcinoma development during follow-up. Hepatology. 2000;31:846-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Saeki A, Tamura S, Ito N, Kiso S, Matsuda Y, Yabuuchi I, Kawata S, Matsuzawa Y. Lack of frameshift mutations at coding mononucleotide repeats in hepatocellular carcinoma in Japanese patients. Cancer. 2000;88:1025-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Sood AK, Buller RE. Genomic instability in ovarian cancer: a reassessment using an arbitrarily primed polymerase chain reaction. Oncogene. 1996;13:2499-2504. [PubMed] |
| 16. | Berg KD, Glaser CL, Thompson RE, Hamilton SR, Griffin CA, Eshleman JR. Detection of microsatellite instability by fluorescence multiplex polymerase chain reaction. J Mol Diagn. 2000;2:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 154] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 18. | Wong N, Lai P, Lee SW, Fan S, Pang E, Liew CT, Sheng Z, Lau JW, Johnson PJ. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol. 1999;154:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 213] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 19. | Wong N, Lai P, Pang E, Fung LF, Sheng Z, Wong V, Wang W, Hayashi Y, Perlman E, Yuna S. Genomic aberrations in human hepatocellular carcinomas of differing etiologies. Clin Cancer Res. 2000;6:4000-4009. [PubMed] |
| 20. | Leung TH, Wong N, Lai PB, Chan A, To KF, Liew CT, Lau WY, Johnson PJ. Identification of four distinct regions of allelic imbalances on chromosome 1 by the combined comparative genomic hybridization and microsatellite analysis on hepatocellular carcinoma. Mod Pathol. 2002;15:1213-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 21. | Yeh SH, Chen PJ, Lai MY, Chen DS. Allelic loss on chromosomes 4q and 16q in hepatocellular carcinoma: association with elevated alpha-fetoprotein production. Gastroenterology. 1996;110:184-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Chan KL, Lee JM, Guan XY, Fan ST, Ng IO. High-density allelotyping of chromosome 8p in hepatocellular carcinoma and clinicopathologic correlation. Cancer. 2002;94:3179-3185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, Nakamura Y. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992;52:5368-5372. [PubMed] |
| 24. | Fujiwara Y, Ohata H, Kuroki T, Koyama K, Tsuchiya E, Monden M, Nakamura Y. Isolation of a candidate tumor suppressor gene on chromosome 8p21.3-p22 that is homologous to an extracellular domain of the PDGF receptor beta gene. Oncogene. 1995;10:891-895. [PubMed] |
| 25. | Piao Z, Kim NG, Kim H, Park C. Deletion mapping on the short arm of chromosome 8 in hepatocellular carcinoma. Cancer Lett. 1999;138:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Liao C, Zhao M, Song H, Uchida K, Yokoyama KK, Li T. Identification of the gene for a novel liver-related putative tumor suppressor at a high-frequency loss of heterozygosity region of chromosome 8p23 in human hepatocellular carcinoma. Hepatology. 2000;32:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Park WS, Lee JH, Park JY, Jeong SW, Shin MS, Kim HS, Lee SK, Lee SN, Lee SH, Park CG. Genetic analysis of the liver putative tumor suppressor (LPTS) gene in hepatocellular carcinomas. Cancer Lett. 2002;178:199-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Sheu JC, Lin YW, Chou HC, Huang GT, Lee HS, Lin YH, Huang SY, Chen CH, Wang JT, Lee PH. Loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in Taiwan. Br J Cancer. 1999;80:468-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Yumoto Y, Hanafusa T, Hada H, Morita T, Ooguchi S, Shinji N, Mitani T, Hamaya K, Koide N, Tsuji T. Loss of heterozygosity and analysis of mutation of p53 in hepatocellular carcinoma. J Gastroenterol Hepatol. 1995;10:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 51] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Rashid A, Wang JS, Qian GS, Lu BX, Hamilton SR, Groopman JD. Genetic alterations in hepatocellular carcinomas: association between loss of chromosome 4q and p53 gene mutations. Br J Cancer. 1999;80:59-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Zhao X, Li J, He Y, Lan F, Fu L, Guo J, Zhao R, Ye Y, He M, Chong W. A novel growth suppressor gene on chromosome 17p13.3 with a high frequency of mutation in human hepatocellular carcinoma. Cancer Res. 2001;61:7383-7387. [PubMed] |
| 32. | Rabe C, Cheng B, Caselmann WH. Molecular mechanisms of hepatitis B virus-associated liver cancer. Dig Dis. 2001;19:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Becker SA, Zhou YZ, Slagle BL. Frequent loss of chromosome 8p in hepatitis B virus-positive hepatocellular carcinomas from China. Cancer Res. 1996;56:5092-5097. [PubMed] |
| 34. | Okabe H, Ikai I, Matsuo K, Satoh S, Momoi H, Kamikawa T, Katsura N, Nishitai R, Takeyama O, Fukumoto M. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology. 2000;31:1073-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Qin LX, Tang ZY, Sham JS, Ma ZC, Ye SL, Zhou XD, Wu ZQ, Trent JM, Guan XY. The association of chromosome 8p deletion and tumor metastasis in human hepatocellular carcinoma. Cancer Res. 1999;59:5662-5665. [PubMed] |
| 36. | Salvucci M, Lemoine A, Saffroy R, Azoulay D, Lepère B, Gaillard S, Bismuth H, Reynès M, Debuire B. Microsatellite instability in European hepatocellular carcinoma. Oncogene. 1999;18:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Piao Z, Kim H, Malkhosyan S, Park C. Frequent chromosomal instability but no microsatellite instability in hepatocellular carcinomas. Int J Oncol. 2000;17:507-512. [PubMed] |
| 38. | Yamamoto H, Itoh F, Fukushima H, Kaneto H, Sasaki S, Ohmura T, Satoh T, Karino Y, Endo T, Toyota J. Infrequent widespread microsatellite instability in hepatocellular carcinomas. Int J Oncol. 2000;16:543-547. [PubMed] |
| 39. | Macdonald GA, Greenson JK, Saito K, Cherian SP, Appelman HD, Boland CR. Microsatellite instability and loss of heterozygosity at DNA mismatch repair gene loci occurs during hepatic carcinogenesis. Hepatology. 1998;28:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |