Published online May 28, 2005. doi: 10.3748/wjg.v11.i20.3020
Revised: December 11, 2003
Accepted: March 12, 2004
Published online: May 28, 2005
AIM: DNA mismatch repair (MMR) is an important mechanism for maintaining fidelity of genomic DNA. Abnormalities in one or more MMR genes are implicated in the development of many cancers. We investigated the role of expression of MMR genes (hMLH1, hPMS1, hPMS2, GTBP/hMSH6, hMSH2) in hepatocellular carcinogenesis.
METHODS: We evaluated the expression level of MMR genes in 33 hepatocellular carcinoma (HCC) cases using the multiplex reverse transcription (RT) PCR assays, as well as in 16 cases of normal adjacent hepatic tissues. β-actin gene was used as an internal control and calibrator for quantification of gene expression.
RESULTS: Out of the 33 studied cases, 25 were HCV positive and 30 (90.9%) showed reduced expression in one or more of the studied MMR genes. Reduced expression was found in hMSH2 (71.9%), hMLH1 (53.3%), GTBP (51.1%), hPMS2 (33.3%) and hPMS1 (6%). A significant correlation was found between reduced expression of hPMS2 (P = 0.0069) and GTBP (P = 0.0034), hPMS2 and non-cirrhosis (P = 0.0197), hMLH1 and high grade. On the other hand, 57.1%, 50%, 20%, 18.8%, and 6% of the normal tissues distant to tumors showed reduced expression of hMSH2, hMLH1, GTBP, hPMS2, and hPMS1 respectively. Multivariate analysis revealed a significant correlation between the expression level of hMSH2 (P = 0.008), hMLH1 (P = 0.001) and GTBP (P = 0.032) and HCC, between hPMS2, GTBP and HCV-associated HCC (P<0.001, 0.002).
CONCLUSION: Reduced expression of MMR genes seems to play an important role in HCV-associated HCC. hPMS2 is likely involved at an early stage of hepatocarcinogenesis since it was detected in normal adjacent tissues. Reduced expression of hPMS2 provides a growth advantage and stimulates proliferation which encourages malignant transformation in non-cirrhotic HCV-infected patients via acquisition of more genetic damages.
-
Citation: Zekri ARN, Sabry GM, Bahnassy AA, Shalaby KA, Abdel-Wahabh SA, Zakaria S. Mismatch repair genes (
hMLH1 ,hPMS1 ,hPMS2 ,GTBP/hMSH6 ,hMSH2 ) in the pathogenesis of hepatocellular carcinoma. World J Gastroenterol 2005; 11(20): 3020-3026 - URL: https://www.wjgnet.com/1007-9327/full/v11/i20/3020.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i20.3020
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, with the highest frequencies reported in Asia and Africa. In countries endemic for viral hepatitis, HCC represents 20-40% of human cancers and most of the cases in these areas are late complications of chronic viral hepatitis[1]. The major risk factors for HCC development are now well defined including chronic viral hepatitis, alcohol, aflatoxin and metabolic disorders. These factors induce malignant transformation by increasing cellular turnover as a consequence of chronic liver injury, regeneration and cirrhosis[2]. Although advances in the treatment of HCC have recently been achieved, the prognosis of this disease generally remains very poor[3].
Understanding the molecular basis of HCC is very important, since novel therapeutic approaches, which target these molecules, may lead to improvement in the prevention and treatment strategies of HCC. Therefore, identification of molecular markers related to prognosis is a vital target to improve the clinical outcome and for a more effective therapy[4]. Recently, some of the multisteps involved in hepatocarcinogenesis have been elucidated. Activation of oncogenes, inactivation of tumor suppressor genes, overexp-ression of certain growth factors and DNA mismatch repair (MMR) defects may contribute to the development of HCC[5].
The DNA MMR system is expressed in all tissues at various levels and plays an important role in the maintenance of genomic integrity as it corrects replicative mismatches of the escaped DNA polymerase proof reading[6]. Biochemical and genetic studies in eukaryotes have defined at least five genes (MSH2, MSH3, GTBP, MLH1, and PMS2) whose protein products are required for DNA MMR[7]. Direct evidence for the association of genetic instability and mutant MMR genes is derived from the biochemical studies in vitro in which nuclear extracts from human tumor cell lines with mutated MMR genes were unable to efficiently repair heter-oduplex DNA fragments[8]. Hence, it was proposed that cells with defective MMR mechanisms had a reduction in the fidelity of DNA and could not correct genetic errors that occurred during cellular replication[9].
Several studies have focused on the impact of defective MMR genes on the pathogenesis of tumors and genes encoding components of the MMR system have been mentioned in relation to several human solid tumors[10-13]. It was shown that defects in MMR genes could lead to a genome-wide instability of the microsatellites[10]. When this occurs in oncogenes or tumor suppressor genes, loss of control over cell growth and proliferation may develop[14]. Moreover, loss of expression of the MMR genes leads to resistance of tumor cells to the damage induced by some chemotherapeutic agents such as 6-thioguanine, some methylating agents, cisplatin and carboplatin[6,15,16]. This acquired resistance could be achieved through several mechanisms including failure to recognize DNA adducts formed by some chemotherapeutic agents or failure to activate signaling pathways that trigger apoptosis[6].
This work was conducted to investigate the possible role of MMR gene defects in the development of HCV-associated HCC through studying the expression levels of hMSH2, GTBP, hMLH1, hPMS-1 and hPMS2 genes using the RT-PCR assay.
In the present study, 49 samples were analyzed (17 samples of HCC and 32 samples of HCC and their associated normal hepatic tissues obtained from morphologically normal areas distant to the tumor in the same patient). All samples were obtained from patients undergoing hepatectomy at the National Cancer Institute, Cairo University during the period 2000-2002. The mean age of patients was 55.77±11.53 years (range 10-80 years) and the male:female ratio was 1.13:1. Out of the 33 HCC cases, 15 were cirrhotic and 18 showed no evidence of cirrhosis. All clinicopathologic features of the patients are illustrated in Table 1. Fresh tumor and normal distant hepatic tissues (NDHTs) were obtained at the operation theater and immediately divided into two pieces. One was snap frozen in liquid nitrogen and stored at -80 °C for subsequent DNA and RNA extraction, the other was fixed in neutral buffered formalin and processed for histopathologic examination to determine tumor type, grade, presence of cirrhosis and percent of neoplastic cells in tumor samples. Only cases composed 75% or more neoplastic cells were included in the study to avoid the neutralizing effect of normal cells. NDHTs were obtained from areas distant to tumors (4-5 cm from the periphery of the tumor) and confirmed to be normal by microscopic examination of hematoxylin and eosin-stained sections. Both normal and neoplastic tissues were examined by two independent pathologists. Unintentional bias was prevented by coding patient tissue samples, so that genomic studies were done without the knowledge of the patient and tumor characteristics. All samples included in this study were HBV-PCR negative.
| Clincopathological features | No. | GTBP | hPMS1 | hPMS2 | hMLH1 | hMSH2 | HCV-RT-PCR | HBV-PCR |
| Age (yr) <50 | 9 | 6 | 0 | 5 | 5 | 7 | 6 | 0 |
| >50 | 24 | 11 | 2 | 6 | 11 | 16 | 19 | 0 |
| Gender M | 18 | 8 | 2 | 4 | 9 | 13 | 12 | 0 |
| F | 15 | 9 | 0 | 7 | 7 | 10 | 13 | 0 |
| Grade I and II | 24 | 12 | 2 | 8 | 15 | 16 | 17 | 0 |
| III | 9 | 5 | 0 | 3 | 1 | 7 | 8 | 0 |
| CirrhosisPresent | 15 | 7 | 1 | 2 | 8 | 12 | 14 | 0 |
| Absent | 18 | 9 | 1 | 9 | 8 | 11 | 11 | 0 |
| Total | 33 | 17 | 2 | 11 | 16 | 23 | 25 | 0 |
A written consent was obtained from all studied patients and the Ethical Committee of the NCI approved the protocol according to the ethical guidelines of the 1975 Declaration of Helsinki.
Sera collected from 5 mL of coagulated blood were aliquoted and stored at -80 °C. All sera from patients and controls were tested for HCV antibody and HBsAg by the third generation ELISA using kits from Innogenetics (Belgium) and Equipar (Saronno, Italy). They were also tested for HBs-Ab, anti-HBc total, anti-HbeAg and HBeAg by the third generation ELISA using kits from Innogenetics (Belgium). All tests were done according to the manufacturer’s instructions.
High molecular weight DNA was prepared from 0.5-2.0 g fresh tissue samples according to standard protocols[17].
RNA from both tumor and normal tissues was extracted using a SV total RNA isolation system (Promega Biotech). The extracted total RNA was assessed for degradation, purity and DNA contamination by a spectrophotometer and electrop-horesis in an ethidium bromide-stained 1.0% agarose gel.
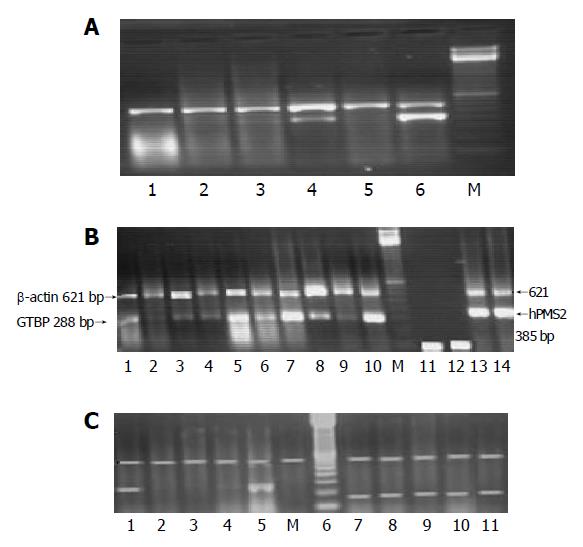
Ten samples of normal human DNA and RNA were extracted from PBL and used to optimize the best conditions for the multiplex PCR of β-actin gene vs each of the five studied MMR genes. Amplification of β-actin gene (621-bp fragments) was performed to test for the presence of artifacts and to set a base line for each sample that enabled the evaluation of the expression of target genes in the multiplex RT-PCR (i.e., semi-quantitation). The β-actin gene fragments were used to monitor DNA contamination. Since all genes were amplified in the same test tube, designing only one pair of primers should be sufficient for controlling DNA contamination. A water control tube containing all reagents except c-DNA was also conducted in each batch of PCR assays to monitor contamination of genomic DNA in the PCR reagents, negative RT-PCR control was used against each sample.
Reverse transcription (RT) of the isolated total RNA was done in 25 μL reaction volume containing 1 volume of Superscript II RT enzyme (Gibco-BRL, Gaithersburg, MD, USA), 2.5 μL 10×RT-buffer [250 mmol/L Tris-HCl pH 8.3, 375 mmol/L KCl, 15 mmol/L MgCl2], 0.251 μL of 100 mmol/L dithiothreitol, 1 μL of 25 ng from random primer, 1.5 μL of 10 mmol/L deoxynucleotide triphosphates, 0.5 μL RNAsin (Promega, USA), 5.0 μL of extracted RNA. Samples were then incubated at 50 °C for 60 min followed by a 4 °C until the PCR amplification reaction.
The PCR and quantitation were performed in a 50 μL reaction volume according to Ref.[12] with some modifications. Briefly, 5 μL of the RT reaction mixture (c-DNA), 2.5 units Taq polymerase (Gibco-BRL, Gaithersburg, MD, USA), 1×PCR buffer [500 mmol/L KCl, 200 mmol/L Tris-HCl, 1.5 mmol/L MgCl2, 1 mg/mL bovine serum albumin], 200 μmol/L each of the deoxyribonucleotide triphosphate and 0.25 μmol/L of each primer were mixed. Samples were denatured at 95 °C for 5 min and subjected to 35 rounds of thermal cycling (T-gradient, Biometra, Germany). Each cycle consisted of denaturation for 1 min at 95 °C, annealing for 1 min at different annealing temperatures as shown in Table 2 and extension for 1 min and 30 s at 72 °C. Samples were then incubated at 72 °C for 10 min. All samples were analyzed twice for MMR by RT-PCR on different days with different RT-PCR mixtures to ensure the reproducibility of results.
| Gene sequence | Size (bp) | Temperature (°C) |
| β-actin | ||
| 5'-ACA CTG TGC CCA ACG AGG-3’ | 621 | 55-59 |
| 5'-AGG GGC CGG TCA T AC T-3’ | ||
| hMPS-2 | ||
| 5'-TGC ATG CAG GAT TTG GAA A-3’ | 385 | 55 |
| 5'-GAA CCC CTC AGA ATC CAC GGA-3’ | ||
| GTBP | ||
| 5'-CCC TCA GCC ACC AAA GAA GCA-3’ | 288 | 56 |
| 5'-CTG CCA CCA CTT CCT CAT CCC-3’ | ||
| hMLH-1 | ||
| 5'-GTG CTG GCA ATC AGG GAC CC-3’ | 215 | 58 |
| 5'-CAC GGT TGA GGC ATT GGG TAG-3’ | ||
| hMPS-1 | ||
| 5'-GCG GCA ACA GTT CGA CTC CTT-3’ | 174 | 57 |
| 5'-AGC CTT GAT ACC CTC CCC GTT-3’ | ||
| hMSH-2 | ||
| 5'-GTC GGC TTC GTG CGC TTC TTT-3’ | 429 | 58 |
| 5'-TCT CTG GCC ATC AAC TGC-3’ HCV | ||
| RB6A | ||
| 5'-GTG AGG AAC TAC TGT CTT CAC G-3 | 266 | 55 |
| RB6B | ||
| 5'-ACT CGC AAG CAC CCT ATC AGG-3’ | ||
| HBV | ||
| LBL | ||
| 5'-CGG ATC CGT GGA GTT ACT CTG | 460 | 55 |
| GTT TTT GC-3’ | ||
| RBL | ||
| 5'-GCA AGC TCT AAC AAC AGT AGT TTC CCG G-3’ |
Fifteen microliters of each PCR product was separated by electrophoresis through a 2.0% ethidium bromide-stained agarose gel and visualized with ultraviolet light. Gels were video-photographed. Then the bands on the photograph were scanned as digital peaks, and the areas of the peaks were calculated in arbitrary units with a digital imaging system (Model IS-1000; Alpha Innotech Co., San Leandro, CA, USA). To evaluate the relative expression levels of target genes in the multiplex RT-PCR, the value of the internal standard (β-actin) in each reaction was used as a normalizing factor and a relative value was calculated for each target genes amplified in the reaction. Reduced expression in any of the studied gene was considered if there was a complete absence or decrease in the intensity of the desired band more than 75% compared to the band of β-actin gene expression which was used as a calibrator. Samples were assayed in batches including both cases and controls. The absence of bands was verified by repeating the multiplex PCR and consistent presence of β-actin gene amplification (Figures 1A-1C).
The RT and PCR were performed as described in Refs.[18] and[19]. After completion of the reaction, 10 μL of each sample was analyzed by electrophoresis through 1.2% ethidium bromide-stained agarose gel.
PCR amplification of HBV was done as previously described in Ref.[20], and after the completion of the reaction, 10 μL of each reaction product was analyzed by electrop-horesis through an ethidium bromide-stained 2% agarose gel.
The results were analyzed using the Graph pad prism computer program (Graph pad software, San Diego, USA). χ2, t-test, and simple correlation tests were used to test for the correlation between the studied parameters. P<0.05 was considered statistically significant.
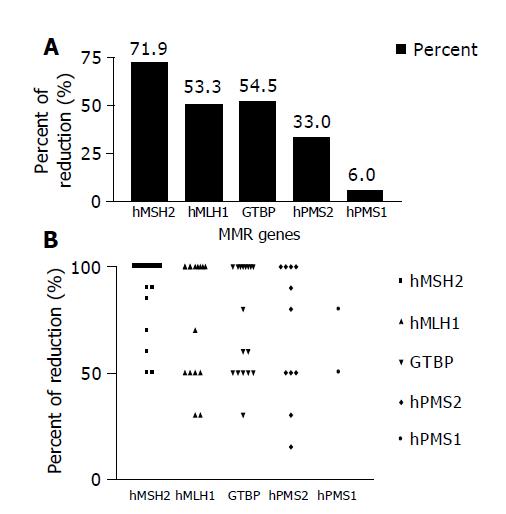
Reduction in the expression of one or more of the five studied MMR genes (hMSH2, hMLH1, GTBP/hMSH6, hPMS2 and hPMS1) was reported in 30/33 (90.9%) HCC cases, whereas none of the cases showed simultaneous reduction in the five genes. The most commonly affected gene was hMSH2, where reduced expression was detected in 23/30 (76.7%) cases, followed by hMLH1 16/30 (53.3%) cases, GTBP 17/30 (56.7%) cases, hPMS2 11/30 (36.7%) cases and hPMS1 2/30 (6.7%) cases (Figures 2A and 2B).
Out of the 30 HCC cases showing reduced expression of one or more genes, 13 (43.3%) had a reduced expression in both hMSH2 and hMLH1, another 13 (43.3%) in both hMSH2 and GTBP, 9 (30%) in both hMLH1 and GTBP, 8 (26.7%) in both GTBP and hPMS2, 6 (20%) in both hMSH2 and hPMS2, another 6 (20%) in both hMLH1 and hPMS2, 2 (6.6%) in both hMSH2 and hPMS1, 1 (3%) in both hMLH1 and hPMS1, and another one (3%) showed a reduction in both GTBP and hPMS1. None of the cases showed a reduction in both hPMS2 and hPMS1. A significant association was found between reduced expressions of GTBP and hPMS2 (P = 0.0034) as well as between hMSH2 and hMLH1 (P = 0.0308, Table 3).
| MMR | hMSH2 | hPMS2 | GTBP | hPMS1 |
| hMLH1 | 13/29 | 6/30 | 9/30 | 1/30 |
| P = 0.0308a | P = 0.178 | P = 0.39 | P = 0.5 | |
| hPMS1 | 2/32 | 0/33 | 1/30 | |
| P = 0.407 | P = 0.391 | P = 0.95 | ||
| GTBP | ||||
| 13/30 | 8/32 | |||
| P = 0.149 | P = 0.0034a | |||
| hPMS2 | 6/32 | |||
| P = 0.407 |
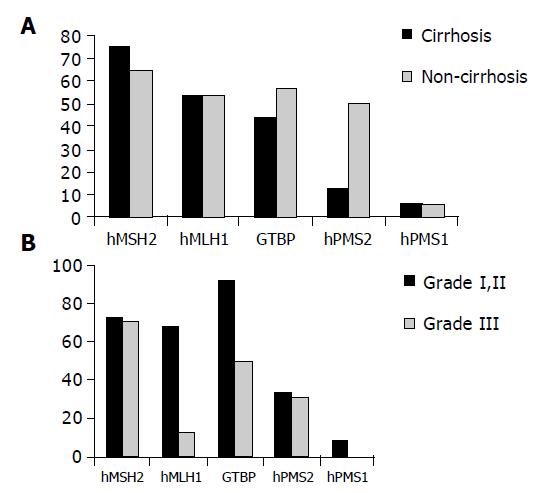
Reduced expression of hMSH2 was found in 12/15 (75%) patients with cirrhosis compared to 11/18 (61.1%) non-cirrhotic cases as well as in 8/15 (53.3%) compared to 8/18 (44.4%) cases for hMLH1, 7/15 (46.7%) compared to 9/18 (50%) cases for GTBP, 2/15 (13.3%) compared to 9/18 (50%) cases for hPMS2 and in 1/15 (6.7%) compared to 1/18 (5.5%) cases for hPMS1. No significant relation was found between reduced expression of any of the studied genes and the clinicopathological features of patients except for the relationship between reduced expression of hPMS2 and non-cirrhotic HCC cases only (P = 0.0197, Figure 3A).
Regarding the relationship between the reduced expression of MMR and grade I and II vs grade III, strong significant relation was found between reduced expression of hMLH1 and grade II (P = 0.0069, Figure 3B).
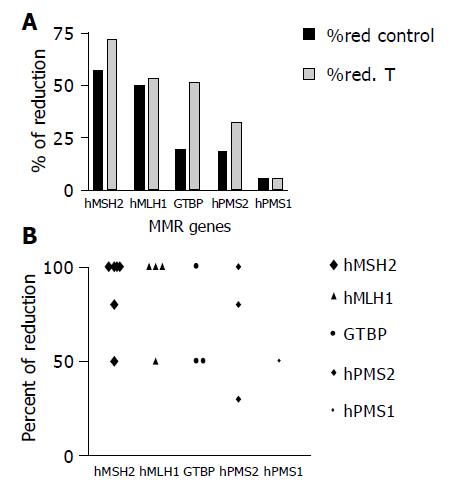
The expression level of hMSH2 was reduced in 8/16 (50%) NDHT samples compared to 23/30 (71.9%) for their associated HCC samples, 7/16 (43.75%) NDHT compared to 16/30 (53.3%) of HCC cases for hMLH1, 3/16 (18.8%) NDHT samples compared to 17/30 (561.7%) of HCC cases for GTBP, in 3/16 (18.8%) NDHT samples compared to 11/30 (36.7%) of HCC cases for hPMS2 and in 1/16 (6.3%) NDHT samples compared to 2/30 (6.6%) of HCC cases for hPMS1. A statistically significant difference in the expression levels of GTBP was found between cases of HCC and NDHT (P = 0.04, Figures 4A and 4B).
A significant relation was found between reduced expression of hPMS2, GTBP and the presence of HCV infection in HCC cases (P<0.001 and 0.002, respectively).
DNA MMR is an important mechanism involved in maintaining the fidelity of genomic DNA. Defective MMR was implicated in a variety of gastrointestinal tumors; however, its role in HCC has been incompletely defined[9]. In this preliminary study, multiplex RT-PCR assay was used to assess the expression levels of five MMR genes in HCV-associated HCCs and their NDHTs.
Reduced expression of hMSH2, hMLH1, GTBP and hPMS2 but not hPMS1 was frequently observed in HCC cases. In general, reduced expression of more than one gene was more frequent than in a single gene. The most frequently affected genes in the present study were MSH2 (71.9%), GTBP (56.7%), and MLH1 (53.3%). The reduced expression could be speculative for somatic mutations affecting these genes in particular and might lead to reduced expression. Literature review showed only very few reports with controversial data in this context. In 1999, Yanno et al[21], reported the presence of mutations in hMSH2 gene in HCC patients and demonstrated that these mutations were closely correlated with the overall survival of patients. In a recent study on the gene expression profile of HCC cases using the cDNA array, reduced expression of MSH2 was found to be a frequent event in HCC and was closely associated with tumor metastasis. In contrast, Wang et al[22], using immunohistochemical techniques did not find any correlation between the expression level of hMLH1 or hMSH2 and the incidence or prognosis of HCC patients. They concluded that defects in hMSH2, hMLH1 did not contribute significantly to hepatocellular carcinogenesis. This controversy in the results of different studies may be attributed to the difference in the detection method of MMR gene defects, the presence of different HCV genotypes of viral infection, epigenetic factors or other factors including race, geographical area, genetic profile and other environmental hazards. It was shown that some mutations could cause premature translation termination that mediates degradation of mutant mRNAs. Moreover, hypermethylation might be another mechanism of gene silencing where hypermethylated genes have reduced mRNA levels. It is also likely that inactivation of transcription factors may reduce the expression of MMR genes with the occurrence of gradual shutdown as a tumor progresses[12].
In addition, several studies demonstrated that the affected MMR genes and the pattern of defects differed according to the type of studied tumors[23]. Whereas in the current study the most frequently affected genes were MSH2, GTBP, and MLH1. Thomas et al[24], studied germline mutations of the MMR genes in African Americans with CRC and found an association between germline mutations of hMLH1 and hMSH2 and the development of CRC in African Americans. Therefore, it seems possible that defects in MMR genes could be induced by the interplay between several genetic and non-genetic factors and even in the same tumor type we may find different patterns of gene defects.
A novel finding in this work is the frequent involvement of GTBP since this gene has not been mentioned before in relation to hepatocellular carcinogenesis. Moreover, in the majority of cases which had a reduced expression of more than one gene, GTBP was usually a partner. Therefore, we assume that genetic defects leading to loss of function of GTBP might represent an important step in the chain of MMR gene defects involved in hepatocyte transformation especially in the presence of HCV infection. This assumption has been confirmed in the present study by finding a significant difference between the expression level of GTBP in tumor and normal hepatic tissues obtained from the same patients. Additional evidence is provided by the high association reported in the present study between reduced expression of GTBP and the presence of HCV in HCC cases.
On the other hand, hMLH1 was the third most frequently affected gene and commonly associated with reduced expression of MSH2. Moreover, reduced expression of hMLH1 in NHTDT was comparable to that reported in tumor tissues in cases from which paired samples of normal and neoplastic tissues were taken (43.2% vs 53.3%). These findings indicate that hMLH1 is a likely candidate gene in hepatocarcinogenesis and possibly exerts its role in the early steps of hepatocyte transformation. Our data in this regard have confirmed the results of Macdonald et al[9], who detected LOH at hMSH2 and/or hMLH1 in malignant, premalignant and adjacent normal hepatic tissues. The high association reported in the current study between reduced expression of hMSH2 and hMLH1 (P = 0.03) suggests the presence of a co-operation between these two genes at a certain stage of hepatocarcinogenesis. Moreover, there was a significant association between reduced expressions of PMS2 gene and absence of cirrhosis in HCC patients (P = 0.0197) since reduced expression of PMS2 was secondary to MSH2 in this group of patients. This provides a possible pathogenetic pathway for the development of HCC in non-cirrhotic patients via failure to repair damaged DNA. Subsequently, it could be proposed that defects in hPMS2 are likely associated with growth advantage and proliferative stimulation which in the absence of effective DNA repair mechanisms may lead to malignant changes in the non-cirrhotic patients. The significant association between reduced expressions of GTBP and hPMS2 in HCC especially in non-cirrhotic cases (P = 0.003) could be explained by the presence of a co-operation between these two genes in the development of HCC.
The significant association reported in the present study between the presence of HCV in HCC cases and reduced expression of hPMS2 and/or GTBP (P<0.001 and 0.002 respectively) suggests that these genes could be targets for HCV in the genetic cascade controlling HCV-associated hepatocarcinogenesis. Alternatively reduced expression or inactivation of these genes may interfere with (prevent) efficient repair of DNA as a result of HCV infection.
It is now well known that HCV-associated HCC involves alterations in the concerned action of proto-oncogenes, growth factors and tumor suppressor genes. The presence of two nuclear localization signals and a DNA binding motif in the HCV core protein suggests a possible functional role for HCV as a gene regulatory element[25]. Moreover, some studies suggested that this protein could interact with certain cellular proto-oncogenes at the transcriptional level, resulting in the promotion of cell proliferation which in the presence of DNA damage and/or in the absence of efficient DNA repair mechanisms affects normal hepatocyte growth and differentiation. Therefore, the pathogenesis of HCV might contribute at least in part to the upregulation of hepatocyte growth induced by HCV core protein and the loss of DNA repair[26].
To the best of our knowledge, no previous study has revealed the relation between MMR defects and HCV infection in the process of hepatocarcinogenesis. Numerous studies have shown that overexpression of growth factor receptors is associated with altered cellular response to damage and DNA repair. In breast and ovarian cancer cells, overexpression of c-erbB2 gene products increased sensitivity to drugs through inhibition of DNA repair[27-29] and we previously reported overexpression of c-erbB2 in HCC and chronic active hepatitis patients in association with HCV genotype-4 which is the predominant genotype in Egyptian patients[25]. It has been clearly indicated that modulation of c-erbB2 could inhibit DNA repair either directly or indirectly[30]. Finally, the high mutation rate occurring in case of MMR gene defects in the coding or regulatory sequences of other genes could lead to more and more genomic damages with an increased probability for neoplastic transformation[6]. This finding may in part explain the high resistance of HCC to the most known regimen of chemotherapy, since it has been previously reported that a close correlation exists between MMR gene defect and resistance to chemotherapy.
In conclusion, the present study represents a step forward for understanding the genetic events that induce HCC in HCV-infected patients. Reduced expression of MSH2, GTBP, MLH1, and PMS2 is a frequent event in HCC. Both GTBP and PMS2 are possible candidates for HCC in HCV-infected patients. However, the mechanisms involved in this process have to be clarified. Reduced expression of hMLH1 occurs in the early stages of hepatocarcinogenesis as evidenced by the finding that there is no significant difference in the expression level of hMLH1 between tumors and NDHTs, and reduced expression of hPMS2 most frequently in non-cirrhotic HCC is associated with HCV infection.
| 1. | Natoli G, Ianni A, Costanzo A, De Petrillo G, Ilari I, Chirillo P, Balsano C, Levrero M. Resistance to Fas-mediated apoptosis in human hepatoma cells. Oncogene. 1995;11:1157-1164. [PubMed] [Cited in This Article: ] |
| 2. | Moradpour D, Blum HE. Molecular aspects of hepatocellular carcinoma. Zentralbl Chir. 2000;125:592-596. [PubMed] [Cited in This Article: ] |
| 3. | William H, Daphne C. Biochemistry and Molecular Biology. Second edition (P. 319- 336). Printed in the USA. 2001;. [Cited in This Article: ] |
| 4. | John M, Peter M, Howley A. The Molecular Basis of cancer. Second edition (P: 423 –428). Printed in the USA. 2001;. [Cited in This Article: ] |
| 5. | Blum HE. Molecular targets for prevention of hepatocellular carcinoma. Dig Dis. 2002;20:81-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Fink D, Nebel S, Norris PS, Aebi S, Kim HK, Haas M, Howell SB. The effect of different chemotherapeutic agents on the enrichment of DNA mismatch repair-deficient tumour cells. Br J Cancer. 1998;77:703-708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 52] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433-1442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 475] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 8. | Risinger JI, Barrett JC, Watson P, Lynch HT, Boyd J. Molecular genetic evidence of the occurrence of breast cancer as an integral tumor in patients with the hereditary nonpolyposis colorectal carcinoma syndrome. Cancer. 1996;77:1836-1843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 9. | Macdonald GA, Greenson JK, Saito K, Cherian SP, Appelman HD, Boland CR. Microsatellite instability and loss of heterozygosity at DNA mismatch repair gene loci occurs during hepatic carcinogenesis. Hepatology. 1998;28:90-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D. Frequent loss of heterozygosity at the DNA mismatch-repair loci hMLH1 and hMSH3 in sporadic breast cancer. Br J Cancer. 1999;79:1012-1017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Han HJ, Yanagisawa A, Kato Y, Park JG, Nakamura Y. Genetic instability in pancreatic cancer and poorly differentiated type of gastric cancer. Cancer Res. 1993;53:5087-5089. [PubMed] [Cited in This Article: ] |
| 12. | Wei Q, Bondy ML, Mao L, Gaun Y, Cheng L, Cunningham J, Fan Y, Bruner JM, Yung WK, Levin VA. Reduced expression of mismatch repair genes measured by multiplex reverse transcription-polymerase chain reaction in human gliomas. Cancer Res. 1997;57:1673-1677. [PubMed] [Cited in This Article: ] |
| 13. | Zekri AR, Bahnassi AA, Bove B, Huang Y, Russo IH, Rogatko A, Shaarawy S, Shawki OA, Hamza MR, Omer S. Allelic instability as a predictor of survival in Egyptian breast cancer patients. Int J Oncol. 1999;15:757-767. [PubMed] [Cited in This Article: ] |
| 14. | Leach FS, Polyak K, Burrell M, Johnson KA, Hill D, Dunlop MG, Wyllie AH, Peltomaki P, de la Chapelle A, Hamilton SR. Expression of the human mismatch repair gene hMSH2 in normal and neoplastic tissues. Cancer Res. 1996;56:235-240. [PubMed] [Cited in This Article: ] |
| 15. | Griffin S, Branch P, Xu YZ, Karran P. DNA mismatch binding and incision at modified guanine bases by extracts of mammalian cells: implications for tolerance to DNA methylation damage. Biochemistry. 1994;33:4787-4793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 82] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 16. | Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90:6424-6428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 304] [Cited by in F6Publishing: 324] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 17. | Sambrook J, Ritsch E, Maniatis T. Molecular cloning. A laboratory manual (2nd edition) Cold Spring Harbor, NY. 1989;. [Cited in This Article: ] |
| 18. | Zekri ARN, Bahnassy AA, Khaled HM, Mansour O, Attia MA. Comparative analysis of different PCR techniques for detection of HCV in hepatocellular carcioma patients. Cancer J. 1995;8:331-335. [Cited in This Article: ] |
| 19. | Zekri AR, Sedkey L, el-Din HM, Abdel-Aziz AO, Viazov S. The pattern of transmission transfusion virus infection in Egyptian patients. Int J Infect Dis. 2002;6:329-331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495-503. [PubMed] [Cited in This Article: ] |
| 21. | Yano M, Asahara T, Dohi K, Mizuno T, Iwamoto KS, Seyama T. Close correlation between a p53 or hMSH2 gene mutation in the tumor and survival of hepatocellular carcinoma patients. Int J Oncol. 1999;14:447-451. [PubMed] [Cited in This Article: ] |
| 22. | Wang L, Bani-Hani A, Montoya DP, Roche PC, Thibodeau SN, Burgart LJ, Roberts LR. hMLH1 and hMSH2 expression in human hepatocellular carcinoma. Int J Oncol. 2001;19:567-570. [PubMed] [Cited in This Article: ] |
| 23. | Heinimann K, Scott RJ, Chappuis P, Weber W, Müller H, Dobbie Z, Hutter P. N-acetyltransferase 2 influences cancer prevalence in hMLH1/hMSH2 mutation carriers. Cancer Res. 1999;59:3038-3040. [PubMed] [Cited in This Article: ] |
| 24. | Weber TK, Chin HM, Rodriguez-Bigas M, Keitz B, Gilligan R, O'Malley L, Urf E, Diba N, Pazik J, Petrelli NJ. Novel hMLH1 and hMSH2 germline mutations in African Americans with colorectal cancer. JAMA. 1999;281:2316-2320. [PubMed] [Cited in This Article: ] |
| 25. | Arteaga CL, Winnier AR, Poirier MC, Lopez-Larraza DM, Shawver LK, Hurd SD, Stewart SJ. p185c-erbB-2 signal enhances cisplatin-induced cytotoxicity in human breast carcinoma cells: association between an oncogenic receptor tyrosine kinase and drug-induced DNA repair. Cancer Res. 1994;54:3758-3765. [PubMed] [Cited in This Article: ] |
| 26. | Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438-4443. [PubMed] [Cited in This Article: ] |
| 27. | Zekri AR, Bahnassy AA, Shaarawy SM, Mansour OA, Maduar MA, Khaled HM, El-Ahmadi O. Hepatitis C virus genotyping in relation to neu-oncoprotein overexpression and the development of hepatocellular carcinoma. J Med Microbiol. 2000;49:89-95. [PubMed] [Cited in This Article: ] |
| 28. | Geisler S, Lønning PE, Aas T, Johnsen H, Fluge O, Haugen DF, Lillehaug JR, Akslen LA, Børresen-Dale AL. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61:2505-2512. [PubMed] [Cited in This Article: ] |
| 29. | Pietras RJ, Fendly BM, Chazin VR, Pegram MD, Howell SB, Slamon DJ. Antibody to HER-2/neu receptor blocks DNA repair after cisplatin in human breast and ovarian cancer cells. Oncogene. 1994;9:1829-1838. [PubMed] [Cited in This Article: ] |
| 30. | Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821-1828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1744] [Cited by in F6Publishing: 1714] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 31. | Soliman AS, Bondy ML, Guan Y, El-Badawi S, Mokhtar N, Bayomi S, Raouf AA, Ismail S, McPherson RS, Abdel-Hakim TF. Reduced expression of mismatch repair genes in colorectal cancer patients in Egypt. Int J Oncol. 1998;12:1315-1319. [PubMed] [Cited in This Article: ] |
| 32. | Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehmé A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881-4886. [PubMed] [Cited in This Article: ] |