Published online May 7, 2005. doi: 10.3748/wjg.v11.i17.2557
Revised: June 25, 2004
Accepted: July 17, 2004
Published online: May 7, 2005
AIM: Fatty acid-CoA ligase 4 (FACL4) is an arachidonate-preferring enzyme which has been shown to be up-regulated in human colon cancer tissues and implicated in the colon tumorigenesis. The purpose of this study was to investigate the role of FACL4 in the human hepatocellular carcinoma (HCC) tumorigenesis and the specific signal pathways involved in this process.
METHODS: We investigated the expression and regulation of FACL4 in HCC, adjacent non-tumorous liver tissues, and cell lines.
RESULTS: In HCC patients, we demonstrated that FACL4 gene expression was markedly elevated in the cancerous tissues than in the adjacent non-cancerous liver tissues. In addition, several human hepatoma cell lines, including Hep3B and HepG2, expressed high levels of FACL4. Stable overex-pression of FACL4 knockdown plasmids (small interfering RNA, siRNA) to Hep3B cells significantly decreased FACL4 expression and subsequently limited the cell proliferation. Treatment of Hep3B cells with 8-bromo-cAMP and SB203508 (p38 MAPK inhibitor) significantly suppressed the FACL4 expression.
CONCLUSION: FACL4 is involved in the HCC tumorigenesis and both cAMP and p38 MAPK pathways are associated with the regulation of FACL4 in HCC.
- Citation: Liang YC, Wu CH, Chu JS, Wang CK, Hung LF, Wang YJ, Ho YS, Chang JG, Lin SY. Involvement of fatty acid-CoA ligase 4 in hepatocellular carcinoma growth: Roles of cyclic AMP and p38 mitogen-activated protein kinase. World J Gastroenterol 2005; 11(17): 2557-2563
- URL: https://www.wjgnet.com/1007-9327/full/v11/i17/2557.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i17.2557
Hepatocellular carcinoma (HCC) is one of the most frequent malignancies worldwide, showing the highest prevalence in Asia and Africa. Most symptomatic HCCs have a low surgical respectability and the disease usually progresses rapidly and has very poor prognosis[1]. It is likely that searching novel molecular or genetic mechanisms of HCC tumorigenesis would shed light on the development of more effective therapeutic strategies against it.
Fatty acid-CoA ligase 4 (FACL4) is an arachidonic acid (AA)-preferring isoenzymes of the acyl-CoA ligase family[2]. Production of acyl-CoA by FACL4 is an essential reaction in mammalian cells. FACL catalyzes the ligation of long chain fatty acids with coenzyme A to produce long chain acyl-CoAs. The resulting acyl-CoAs can be subsequently metabolized into the pathways of β-oxidation, glycerolipid synthesis, cholesteryl ester synthesis, desaturation, elongation, and protein acylation and can also serve as signaling molecules[3]. There are five FACL isoenzymes present in human tissues[4,5]. Of these, FACL4 could highly express in human steroidogenic tissues, such as placenta, brain, testis, ovary, spleen, and adrenal cortex, whereas it is less expressed in the gastrointestinal system, including liver[6]. FACL4 is highly elevated in colon adenocarcinoma and plays an important role in colon carcinogenesis[7]. A previous study also demonstrated that exogenous addition of AA induced apoptosis in colon cancer cell lines and so did triacsin C, a FACL4 inhibitor[8], suggesting that FACL4 may be involved in the tumorigenesis process. However, the molecular pathways underlying FACL4-associated tumorigenesis process are still not clear.
Cyclic AMP (cAMP) is a second messenger in eukaryotes, playing a crucial role in the intracellular signal transduction of various stimuli, and controlling a wide variety of cellular events including cell proliferation, differentiation, apoptosis, and several cytoskeletal remodeling functions. cAMP has traditionally been thought to act exclusively through cAMP-dependent protein kinase (PKA), but recent studies have clearly shown that not all effects of cAMP are mediated by a general activation of PKA[9]. A previous study demonstrated that an elevated cAMP could suppress the N-formyl methionyl-leucyl-phenylalanine-induced p38 mitogen-activated protein kinase (MAPK) activation in the hypertonic stress suppression of neutrophil function[10]. Another experiment indicated that cAMP could inhibit multiple signaling pathways, including p38, induced by an amyloid precursor protein in human monocytic cell line THP-1[11]. These findings suggested a link between elevation of cAMP and inhibition of p38 activation. The p38 pathway also plays an essential role in regulating many cellular processes including inflammation, cell differentiation, cell cycle, cell growth and death[12]. Regarding the effect of cAMP on the cell cycle progression, several reports demonstrated that cAMP could induce G1 synchronization, growth arrest, and terminal differentiation in certain cells[13,14]. For example, Lee et al[15], showed that cAMP could induce G1 arrest and apoptosis in hepatoma cells.
In this study, we found that FACL4 was up-regulated in HCC specimens, and knockdown of FACL4 gene expression resulted in growth inhibition of hepatoma cells. In addition, we demonstrated that both cAMP and p38 were involved in the regulation of FACL4. Our results suggest that overexpression of FACL4 might contribute to the growth of hepatoma cells or HCC tumorigenesis.
A total of 16 HCC tissues and their corresponding non-cancerous liver tissues obtained from patients who received surgical treatment were enrolled in this study. Tumors were carefully dissected from adjacent normal tissues and all the malignant and non-malignant tissue specimens were confirmed by pathological examination. This study was approved by the Human Study Review Board of the Taipei Medical University. Hep3B (HB-8064; American Type Culture Collection) originated from a HCC was used in this study.
Total RNA was isolated from human tissue specimens or Hep3B cells, and cDNA was prepared as previously described[16]. The amplification of FACL4 cDNA was performed by incubating 20 ng equivalents of cDNA in 100 mmol/L Tris-HCl buffer, pH 8.3, containing 500 mmol/L KCl, 15 mmol/L MgCl2, 0.1% gelatin, 200 μmol/L concentration of each dNTP, and 50 U/mL Super Taq DNA polymerase with the following oligonucleotide primers: 5’-ATGGCAAA-GAGAATAAAAGCT-3’ and 5’-TGGACTTTGCTC-ATAACATTC-3’. The cDNA sequence of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also amplified as a control in the same method using the following primers: 5’-CCACCCATGGCAAATTCCATGGCA-3’ and 5’-TCTAGACGGCAGGTCAGGTCCACC-3’. Thermal cycle conditions were as follows: 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min, and 1 cycle at 72 °C for 10 min. PCR products were analyzed on 1.0% agarose gels. The size of the RT-PCR products for FACL4 and GAPDH was 1046 and 598 bp, respectively.
Cells were washed with cold PBS, lysed in Golden lysis buffer, and performed Western blotting as described previously[17]. The blotting procedures were first with anti-phosphorylated p38 polyclonal antibody (1:1000 dilution, Cell Signaling Technology, Beverly, MA), anti-p38 polyclonal antibody (1:1000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA), or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1:2000 dilution, Biogenesis, Kingston, NH), and then with anti-mouse or anti-rabbit IgG antibody conjugated to horseradish peroxidase (1:5000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) and visualized bands using enhanced chemiluminescence kits (2 min, ECL, Amersham). The densities of the bands were quantitated with a computer densitometer (IS-1000 Digital Imaging System).
Equal amounts of total cellular protein (200 μg) were immunoprecipitated with p38 specific antibody (Santa Cruz Biotechnology) and protein A/G-PLUS agarose for 15 h at 4 °C. The kinase reactions were carried out in a final volume of 40 μL containing 1 μg of substrate of glutathione S-transferase (Gst)-ATF2 fusion protein (Santa Cruz Biotechnology), 20 μmol/L cold ATP, 5 μCi [r-32p] ATP (5000 Ci/mmol, Amersham) and incubated for 20 min at 25 °C. Each sample was mixed with 10 μL of 5×Laemmli’s loading buffer to stop the reaction, heated for 10 min at 100 °C, and subjected to SDS-PAGE. The gels were dried, visualized by autoradiography, and quantitated by densitometry (IS-1000 Digital Imaging System).
As previously described[19], paraffin-embedded blocks were incubated with anti-phosphorylated p38 antibody according to the manufacturer’s instructions (DAKO LSAB+kit, Dako Corp., Carpinteria, CA).
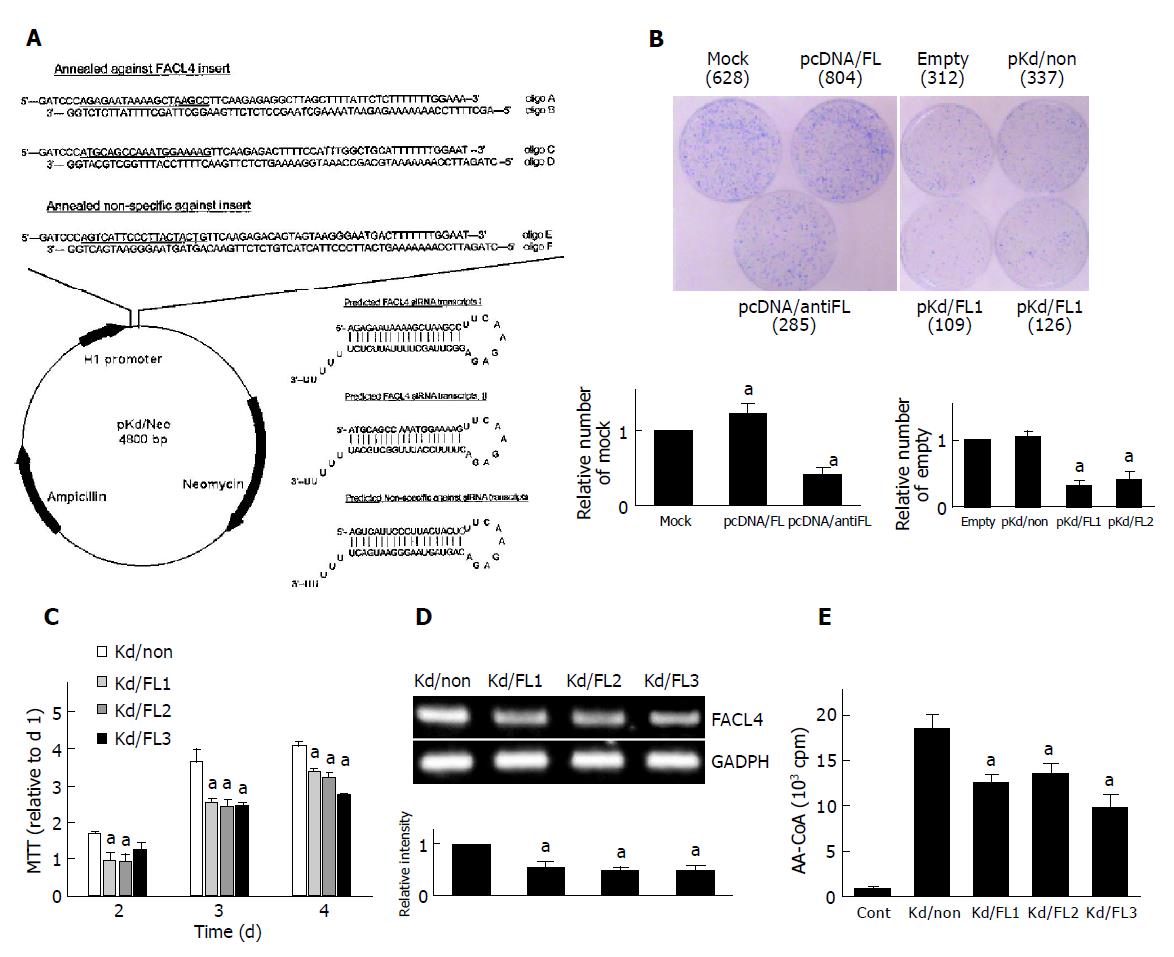
Briefly, the siRNA stable expression vector-pKd/Neo was constructed from pcDNA3 (with CMV promoter and neomycin selectable gene), in which the CMV promoter was replaced by human H1 RNA polymerase III promoter of pSilencer 3.0-H1 vector (Ambion Inc., Worcester, Massachusetts). Therefore, pKd/Neo vector was ready for insertion of specific double-strand oligonucleotides and could stably express siRNA. For knockdown of FACL4, the complementary oligonucleotides A, B, C and D were designed according to the guidelines as described in Ambion website (http://www.ambion.com), containing a 19-mer oligonucleotides (Figure 2A, underlined) corresponding to nucleotides 322-340 and 513-531, respectively, of human FACL4 (GenBank accession no. XM 017658) coding region. Another two randomly-designed oligonucleotides (oligo E and F) lacking FACL4 and other genes (blast with GenBank) were used as a negative control to verify siRNA mechanism. After annealing the complementary oligonucleotides, oligoA/B, oligoC/D, and oligoE/F were inserted into the pKd/Neo vector in specific restriction endonuclease sites (BamHI/XbaI), to generate pKd/FL1, pKd/FL2, and pKd/non-plasmids, respectively.
For selection of stable FACL4 knockdown cell lines, Hep3B cells were transfected with pKd/FL2 or pKd/non plasmid[20] and treated with G418 for 3 wk. Three potential FACL4 knockdown clones (Kd/FL1-3) and one non-specific knockdown clone (Kd/non) were chosen to examine the growth rate. The FACL4 mRNA level of the Kd/non-clone was measured by RT-PCR and showed no significant difference in the parent cells.
Total RNA was isolated from Hep3B cells, and the full length FACL4 cDNA was synthesized by RT-PCR with primers 5’-ACGCTATGGCAAAGAGAATAA-3’ and 5’-AGACAACAACATTTTATTTGC-3’. The RT-PCR products were inserted into the linearized pTARGET vector (Promega) through TA ligation mechanism to obtain pTar/FL plasmid. To construct the FACL4 sense expression plasmid pcDNA/FL, a Kpn I/Xho I fragment excised from the vector pTar/FL was inserted into the pcDNA3.1 vector in the sense orientation. On the other hand, a BamHI/NotI fragment excised from the vector pTar/FL was inserted into the pcDNA3.1 vector in the antisense orientation to obtain pcDNA/antiFL plasmid. Sequence identities were confirmed using an ABI PRISM 377 DNA analysis system (Perkin-Elmer Corp., Taipei, Taiwan).
FACL4 enzyme activity was measured by counting AA-CoA formed from AA as described previously with some modifi-cations[21]. Total protein was extracted by Golden lysis buffer. Fifty micrograms of total protein was incubated with 0.5 μL [14C] AA (50 μCi/mL) substrate in a final volume of 200 μL reaction buffer (0.1 mol/L Tris, pH 7.5, 1 mmol/L GSH, 0.1 mmol/L hydroquinone, 6.7 mmol/L ATP, 20 mmol/L MgCl2, 67 μmol/L CoA) for 5 min. For removing the residual AA (in petroleum ether phase), the reaction mixture was extracted twice with 1.2 mL petroleum ether, and the aqueous phase was then acidified by the addition of 7.5 μL of 1 N HCl to approximately pH 3.0. After two extractions of ethyl acetate, the aqueous phase containing[14C] AA-CoA was collected, mixed with the scintillation cocktail (Cocktail EX2, VWR International Ltd.), and the radioactivities (cpm) were estimated by a liquid scintillator (Beckman L-6000) .
To study the colony formation in monolayer culture, cells were plated at 6×105 per 6-cm dish, and transfected with an individual plasmid as indicated in Figure 2B. After 48 h post-transfection, the cells were treated with G418 and the colonies were stained with Coomassie blue at 4 wk after transfection.
The cells were seeded in a 96-well plate for 1-4 d and the cell number was estimated by a tetrazolium-based semiauto-mated colorimetric assay (MTT assay)[22]. Three clones of FACL4 knockdown cells (Kd/FL1-3) and a negative control clone (Kd/non) were used in this test.
Results were expressed as mean±SE. for each study. Data were analyzed by Student’s t-test and P≤0.05 was considered statistically significant.
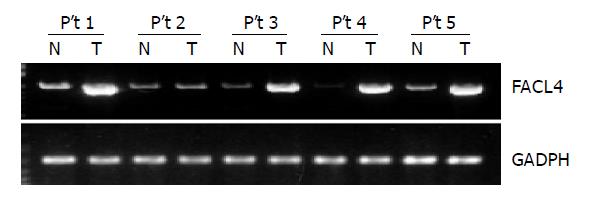
Sixteen HCC tissues and their corresponding non-cancerous tissues from the same patient were used to detect the expression of FACL4 by RT-PCR. As shown in the Figure 1, a representative result from five patients indicated that FACL4 expression was significantly increased in four HCC tissues in comparison with their individual adjacent non-cancerous tissues. Altogether, 13 of 16 (81.3%) HCC cases showed at least two-fold increased expression of FACL4 in cancerous tissues. The change in the expression level of FACL4 relative to non-cancerous tissues ranged from 2.3 to 27.52 folds, the average increase was 8.68 (n = 13). Our findings suggested that FACL4 played an important role in HCC tumorigenesis.
The human HCC cell line Hep3B was used in this study, which expressed high levels of FACL4 as the human colon cancer cell line Colo205. To investigate the role of FACL4 in cell growth, small interfering RNA (siRNA) was used to disrupt the expression of endogenous FACL4. Several studies used siRNA expression plasmids or double-stranded siRNA oligonucleotides to suppress the specific protein expression by transition transfection[23]. However, little is known whether the continuous and stable expression of siRNA also functions well for a long period of time in mammalian cells. In this study, we constructed the FACL4 siRNA expression plasmid (Figure 2A) with neomycin-resistant genes and selected several stable FACL4 siRNA expression clones. During the examination period (about 4 mo), they always expressed a lower level of FACL4 than the parent Hep3B cells. These results indicated that siRNA expression plasmids were able to express continuously for a long period of time without significant toxicity to mammalian cells.
To examine whether suppression of FACL4 could affect HCC cell growth, Hep3B cells were transfected with FACL4 sense and antisense expression plasmids. In a representative experiment, about 628 colonies were found in the cells transfected with mock expression plasmids after selection of G418 for 4 wk. However, about 804 and 285 colonies were found in the cells transfected with pcDNA/FL and pcDNA/antiFL, respectively (Figure 2B, left). Similar results were found in the cells transfected with FACL4 siRNA expression plasmids. Then, Hep3B cells were transfected with two kinds of FACL4 siRNA expression plasmids (pKd/FL1 & pKd/FL2) or one control siRNA expression plasmid (pKd/non). FACL4 knockdown significantly reduced the number of colony formation in comparison with controls. The number of colony formation was about 109, 126, and 337 in the cells transfected with pKd/FL1, pKd/FL2, and pKd/non, respectively (Figure 2B, right). These results indicated that decreased FACL4 expression significantly inhibited the colony formation number and the constitutive expression of FACL4 was associated with the growth of HCC.
Three potential FACL4 knockdown clones (Kd/FL1-3) and a control, non-specific siRNA expression clone (Kd/non) were chosen to compare their growth rate by MTT assay. Three clones (Kd/FL1-3) decreased about 50% in FACL4 expression and their growth rate also decreased in comparison with the Kd/non-clone (Figures 2C and 2D); representative experiments were shown. The decrease of growth rate was not due to the increase of Hep3B cell death by trypan blue exclusion assay. Moreover, FACL4 protein levels were confirmed by FACL4 enzyme activity assay. As shown in Figure 2E, Kd/FL1-3 clones had a lower FACL4 activity as the mRNA level by RT-PCR. These results indicated that the expression levels of FACL4 were associated with cell growth, and FACL4 overexpression might contribute to the tumorigenesis of HCC.
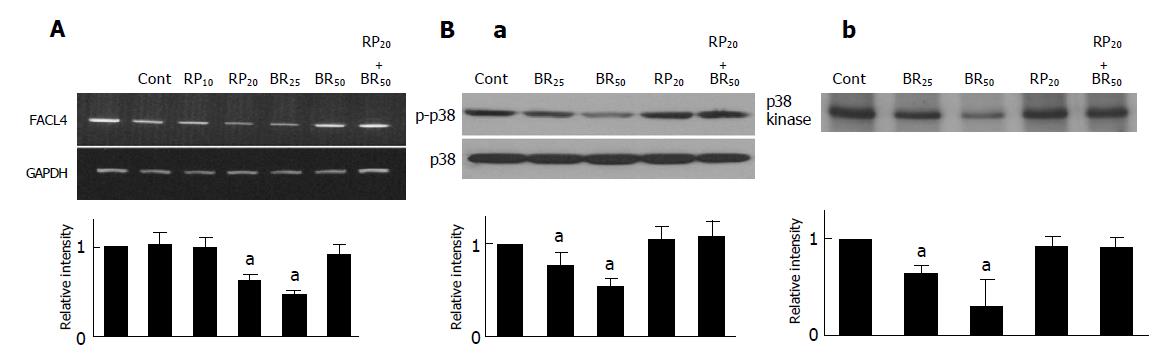
Previous studies demonstrated that elevated cAMP was able to inhibit fatty acid synthesis, and the process was mediated indirectly by cAMP-dependent protein kinase[24]. Therefore, we used 8-bromo-cAMP, a metabolism resistant analog of cAMP, and Rp-adenosine-3’,5’-cyclic monophosphorothioate (Rp-cAMPS), a competitive cAMP antagonist for cAMP-receptor proteins, to examine whether cAMP also participated in the regulation of FACL4 in hepatoma cells. As shown in Figure 3A, treatment of Hep3B cells with 8-bromo-cAMP could dose dependently suppress FACL4 mRNA expression and this effect was reversed by pretreatment with Rp-cAMPS. On the other hand, a PKA inhibitor, H-89 did not affect the expression of FACL4 (data not shown). These results suggested that cAMP negatively regulated FACL4 expression in PKA-independent pathways in Hep3B cells.
To further study which downstream signaling pathways were involved in the regulation of FACL4 by cAMP, we focused on the MAPKs members. Since cAMP could activate the ERK kinase activity and inhibit the p38 kinase activity[10,11,25], we next examined whether cAMP regulated the kinase activity of MAPK members in Hep3B cells. Treatment of Hep3B cells with 8-bromo-cAMP dose dependently suppressed p38 phosphorylation and this effect was reversed by pretreatment with Rp-cAMPS (Figure 3B(a)). To further examine that cAMP not only inhibited p38 phosphorylation but also p38 kinase activity, we performed in vitro kinase activity assay using Gst-ATF2 fusion protein as a substrate. As shown in Figure 3B(b), 8-bromo-cAMP dose-dependently suppressed the p38 kinase activity and this effect was reversed by pretreatment with Rp-cAMPS. However, 8-bromo-cAMP did not affect the phosphorylation and kinase activity of the other MAPK members, ERK and JNK (data not shown). These results suggested that cAMP negatively regulated the p38 kinase activity in Hep3B cells.
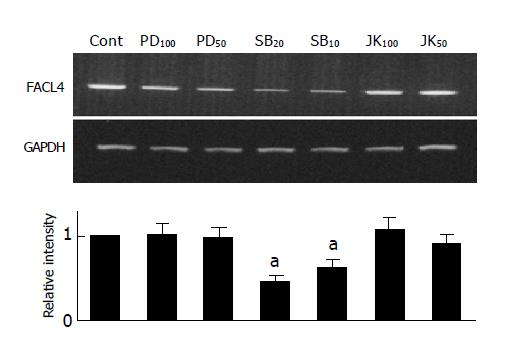
Since elevated cAMP was able to inhibit both of FACL4 expression and p38 activity (Figures 3A and 3B). To examine whether p38 also participated in the regulation of FACL4 gene expression, we used a MEK 1 inhibitor (PD98059), a p38 inhibitor (SB203580) and a JNK inhibitor (SP600125) to examine their effects on the FACL4 expression in Hep3B cells. As shown in Figure 4, treatment of the Hep3B cells with SB203580, but not PD98059 or SP600125, could block the FACL4 mRNA expression, suggesting that p38 but not ERK or JNK was involved in this process. In addition, SB203580 inhibited FACL4 expression in a dose-dependent manner.
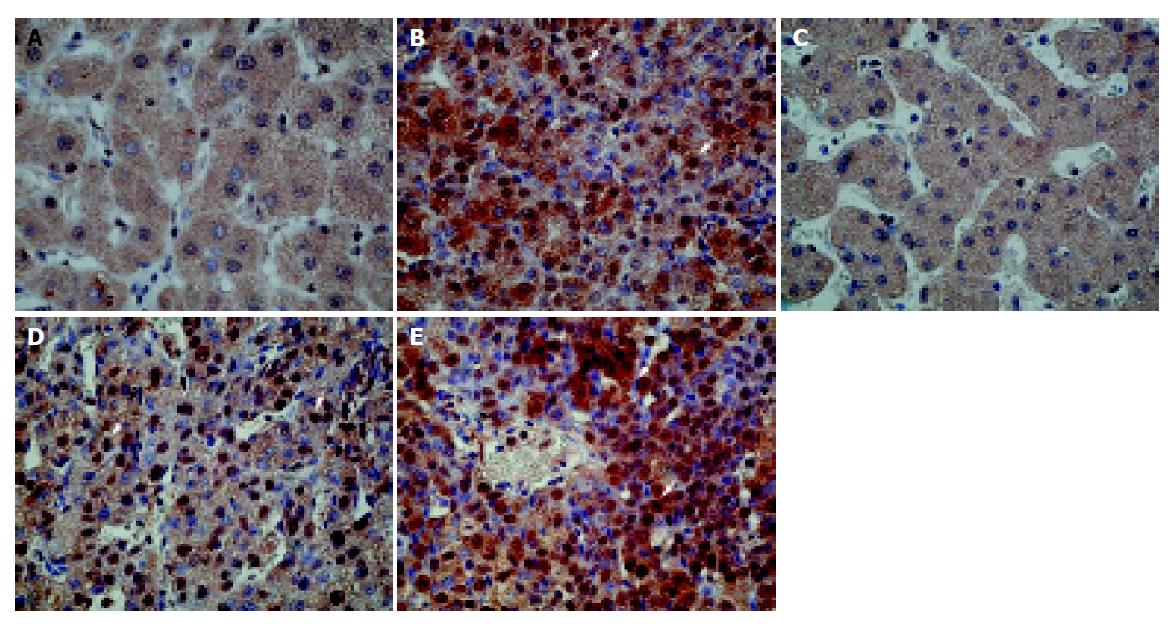
The above-mentioned in vitro experiments demonstrated that FACL4 expression might also mediate through p38 pathway in HCC cell lines. We then tested whether the expression of phosphorylated p38 (p-p38), the activated form of p38, was also elevated in human HCC tissues that could overexpress FACL4. In our 16 HCC tissues, 10 (62.5%) showed an increased p-p38 expression. Importantly, all the p-p38 increased expression cases were in the FACL4 mRNA up-regulated group. Figure 5 shows the immunohistochemistry study of two representative HCC patients. The cancerous tissues strongly expressed p-p38 protein (Figures 5B and 5E). In contrast, p-p38 immunoreactivities were very low in the adjacent non-cancerous tissues (Figures 5A and 5C). Interestingly, when the tumorigenesis process was from the cirrhotic nodules (Figure 5C) to well differentiated (Figure 5D) HCC and then to moderately differentiated (Figure 5E) HCC, the p-p38 immunoreactivities were expressed from low to strong.
A previous report demonstrated that the different siRNA sequences targeting different positions in the specific gene could influence the knockdown efficiency of the specific gene[26]. In this study, we used two kinds of siRNA to knockdown the FACL4 expression and three clones were decreased about 50% in FACL4 mRNA levels and enzyme activity (Figures 2D and 2E). However, the inhibition of FACL4 expression significantly limited HCC cell proliferation and this effect was related to the expression level of FACL4 (Figure 2). It would be of great interest to perform experiments using several different FACL4-specific siRNAs to fully knockdown the gene expression and to examine their impacts on HCC cell proliferation.
The p38 is one of the discrete signaling cascades for diverse extracellular stimuli, and functions to regulate cellular processes. Activation of p38 often through cytokines, growth factors, pathogens and stress environments, could mediate signal transductions into the nuclei to turn on the responsive genes[12]. The upstream activators of p38 are diversified and involved in various signal events including cAMP dependent pathways. One recent study demonstrated that cAMP elevation could inhibit LPS-induced IL-12 production through suppression of p38 in macrophages[27]. These findings are consistent with our results that FACL4 expression was mediated through p38 pathway and elevated cAMP suppressed FACL4 expression by inhibition of p38 activity. A recent report indicated that histone H3 phosphorylation and the activity of chromatin remodeling complexes were regulated by the p38 pathway[28]. It is possible that p38-dependent histone phosphorylation could regulate FACL4 promoters through increasing recruitment of transcription factors. Another report showed the potential transcriptional elements in the promoter region of FACL4 gene[2]. The p38 might regulate the transcriptional activity directly in FACL4 promoters. Further study is required to clarify the transcription factors targeted by the p38 pathways toward the expression of FACL4 in HCC.
In conclusion, our data suggest that p38 and cAMP pathways are involved in the regulation of FACL4 in HCC. Furthermore, FACL4 products induce liver cancer cell proliferation and participate in HCC tumorigenesis. Therefore, development of methods to selectively inhibit FACL4 expression might lead to a novel strategy for the treatment of HCC.
| 1. | Colombo M. Hepatocellular carcinoma. J Hepatol. 1992;15:225-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 157] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Minekura H, Kang MJ, Inagaki Y, Cho YY, Suzuki H, Fujino T, Yamamoto TT. Exon/intron organization and transcription units of the human acyl-CoA synthetase 4 gene. Biochem Biophys Res Commun. 2001;286:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1-12. [PubMed] |
| 4. | Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. Structure and regulation of rat long-chain acyl-CoA synthetase. J Biol Chem. 1990;265:8681-8685. [PubMed] |
| 5. | Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci USA. 1997;94:2880-2884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 210] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 6. | Cao Y, Traer E, Zimmerman GA, McIntyre TM, Prescott SM. Cloning, expression, and chromosomal localization of human long-chain fatty acid-CoA ligase 4 (FACL4). Genomics. 1998;49:327-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 95] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Cao Y, Dave KB, Doan TP, Prescott SM. Fatty acid CoA ligase 4 is up-regulated in colon adenocarcinoma. Cancer Res. 2001;61:8429-8434. [PubMed] |
| 8. | Cao Y, Pearman AT, Zimmerman GA, McIntyre TM, Prescott SM. Intracellular unesterified arachidonic acid signals apoptosis. Proc Natl Acad Sci USA. 2000;97:11280-11285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 316] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Dremier S, Kopperud R, Doskeland SO, Dumont JE, Maenhaut C. Search for new cyclic AMP-binding proteins. FEBS Lett. 2003;546:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Orlic T, Loomis WH, Shreve A, Namiki S, Junger WG. Hypertonicity increases cAMP in PMN and blocks oxidative burst by PKA-dependent and -independent mechanisms. Am J Physiol Cell Physiol. 2002;282:C1261-C1269. [PubMed] |
| 11. | Chong YH, Shin YJ, Suh YH. Cyclic AMP inhibition of tumor necrosis factor alpha production induced by amyloidogenic C-terminal peptide of Alzheimer's amyloid precursor protein in macrophages: involvement of multiple intracellular pathways and cyclic AMP response element binding protein. Mol Pharmacol. 2003;63:690-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1191] [Cited by in RCA: 1244] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 13. | Trepel JB, Colamonici OR, Kelly K, Schwab G, Watt RA, Sausville EA, Jaffe ES, Neckers LM. Transcriptional inactivation of c-myc and the transferrin receptor in dibutyryl cyclic AMP-treated HL-60 cells. Mol Cell Biol. 1987;7:2644-2648. [PubMed] |
| 14. | Bang YJ, Pirnia F, Fang WG, Kang WK, Sartor O, Whitesell L, Ha MJ, Tsokos M, Sheahan MD, Nguyen P. Terminal neuroendocrine differentiation of human prostate carcinoma cells in response to increased intracellular cyclic AMP. Proc Natl Acad Sci USA. 1994;91:5330-5334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 193] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 15. | Lee J, Choi YH, Nguyen P, Kim JS, Lee SJ, Trepel JB. Cyclic AMP induces inhibition of cyclin A expression and growth arrest in human hepatoma cells. Biochim Biophys Acta. 1999;1449:261-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Lin SY, Liang YC, Ho YS, Tsai SH, Pan S, Lee WS. Involvement of both extracellular signal-regulated kinase and c-jun N-terminal kinase pathways in the 12-O-tetradecanoylphorbol-13-acetate-induced upregulation of p21(Cip1) in colon cancer cells. Mol Carcinog. 2002;35:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Lin SY, Chang YT, Liu JD, Yu CH, Ho YS, Lee YH, Lee WS. Molecular mechanisms of apoptosis induced by magnolol in colon and liver cancer cells. Mol Carcinog. 2001;32:73-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Lin SY, Liu JD, Chang HC, Yeh SD, Lin CH, Lee WS. Magnolol suppresses proliferation of cultured human colon and liver cancer cells by inhibiting DNA synthesis and activating apoptosis. J Cell Biochem. 2002;84:532-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 84] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Lee WS, Chen RJ, Wang YJ, Tseng H, Jeng JH, Lin SY, Liang YC, Chen CH, Lin CH, Lin JK. In vitro and in vivo studies of the anticancer action of terbinafine in human cancer cell lines: G0/G1 p53-associated cell cycle arrest. Int J Cancer. 2003;106:125-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Liang YC, Huang YT, Tsai SH, Lin-Shiau SY, Chen CF, Lin JK. Suppression of inducible cyclooxygenase and inducible nitric oxide synthase by apigenin and related flavonoids in mouse macrophages. Carcinogenesis. 1999;20:1945-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 385] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 21. | Sakuma S, Fujimoto Y, Katoh Y, Kitao A, Fujita T. The effects of fatty acyl CoA esters on the formation of prostaglandin and arachidonoyl-CoA formed from arachidonic acid in rabbit kidney medulla microsomes. Prostaglandins Leukot Essent Fatty Acids. 2001;64:61-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 22. | Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38285] [Cited by in RCA: 39373] [Article Influence: 937.5] [Reference Citation Analysis (0)] |
| 23. | Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6971] [Cited by in RCA: 7019] [Article Influence: 292.5] [Reference Citation Analysis (0)] |
| 24. | Cohen P, Hardie DG. The actions of cyclic AMP on biosynthetic processes are mediated indirectly by cyclic AMP-dependent protein kinase. Biochim Biophys Acta. 1991;1094:292-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 705] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 26. | McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1044] [Cited by in RCA: 991] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 27. | Feng WG, Wang YB, Zhang JS, Wang XY, Li CL, Chang ZL. cAMP elevators inhibit LPS-induced IL-12 p40 expression by interfering with phosphorylation of p38 MAPK in murine peritoneal macrophages. Cell Res. 2002;12:331-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 573] [Article Influence: 24.9] [Reference Citation Analysis (0)] |