INTRODUCTION
Hepatitis B virus (HBV) is a small enveloped DNA virus, whose envelope contains three related surface glycoproteins called the large (L), middle (M) and small (S) proteins. All these proteins are the products of a single open reading frame, which is divided into preS1, preS2 and S regions[1]. They arise from three separate translation initiation AUG codons and share a common C-terminal. The S protein (226 amino acid residues) is encoded by the S gene. The M protein contains preS2 (55 amino acid residues) and S regions, while L protein contains preS1 (108 or 119 amino acid residues, depending on the subtype), preS2 and S regions.
HBV is the major etiologic agent for hepatitis B in humans[2]. It may cause acute and chronic hepatitis with serious sequelae such as cirrhosis and hepatocellular carcinoma (HCC)[3]. There are about 400 million HBV carriers worldwide[4] with about one million deaths per year due to HBV-associated liver cancer[5]. Whereas a previous study claimed that mass vaccination was effective in preventing HBV infection and in declining the incidence of HCC[6]. These enormous successes were achieved with plasma-derived vaccines, and later with yeast-derived recombinant vaccines[7,8]. However, both vaccines have a none-response rate of up to 10% in healthy vaccines after immunization with the licensed vaccines containing only the S-protein component of HBV surface antigen (HBsAg) particles[9]. Therefore, it is necessary to develop more effective and highly immunogenic vaccines.
For the development of vaccines against HBV, the preS1 sequence of HBsAg has been shown to be a particularly efficient immunogen at T-and B-cell levels[10,11]. In general, epitopes of proteins should be incorporated into recombinant vaccines for the production of poly-and monoclonal antib-odies[12]. Great deals of studies have shown that there are many immunogenic B-cell epitopes in the preS1 domain. Five distinct antibody-binding sites within the preS1 region of HBsAg/P43 of the adw subtype: preS1 (16-27), preS1 (32-53), preS1 (41-53), preS1 (94-105) and preS1 (106-117) have been determined in mice[13]. A putative B-cell epitope biased to the C-terminal region, preS1 (57-119), was identified in a commercially available immunoglobulin prep-ared from chronically HBV-infected donors[14]. Additionally, many epitopes of monoclonal antibodies directed against preS1 have also been mapped such as preS1 (27-35), preS1 (72-78)[15], preS1 (30-35)[16], preS1 (19-26), preS1 (37-45)[17,18], preS1 (37-43)[19] and preS1 (32-47)[20]. Many T-cell epitopes in preS1 region have also been identified, such as preS1 (21-30), preS1 (29-48)[21], preS1 (12-21) and preS1 (94-117)[13]. On the other hand, the incorporate site and size of preS1 seque-nces are controversial. Some candidate vaccines integrated all preS1 sequences[22-24]; other incorporated partial sequences of preS1 such as preS1 (21-47)[25], preS1 (12-52)[26] and preS1 (1-42)[27]. So it is important to further identify the immunogenic domains in HBsAg preS1 region for the design of vaccines against HBV.
Some reports pointed out that there were no important B-cell epitopes in preS1 (1-20) domains[13,28]. In the present study, eight GST-preS1 fusion proteins overlapped in the preS1 (21-119) region were highly expressed in E.coli and used to identify its immunogenic domains in mice and hum-ans by Western blot analysis and ELISA.
MATERIALS AND METHODS
Expression, purification of GST-preS1 fusion proteins
Eight overlapping fragments, preS1 (21-47), preS1 (34-59), preS1 (48-70), preS1 (60-85), preS1 (71-94), preS1 (86-109), preS1 (95-119) and preS1 (21-119) of HBV adr subtype were individually expressed as a fusion protein with GST. The coding sequences of these preS1 fragments were synthesized by PCR from pADR-1 using 5’- and 3’-primers and subcloned into the BamHI and EcoRI sites of pGEX 4T-1 (Pharmacia) to yield expression plasmids. To express the GST-preS1 fusion proteins, expression plasmids were transformed into E.coli BL21 (gold) cells and the expression of fusion proteins was induced by 0.1 mmol/L isoprophyl-β-D-thiogalactopyranoside (IPTG) at 37 °C for 2 h. The induced cells were harvested and sonicated. Supernatants of the cell lysates were affinity purified by glutathione (GSH)-sepharose 4B (Sigma) column chromatography. The quantity of purified fusion proteins was measured by Lowry’s method using bovine serum albumin as the standard. The procedures in detail were described in a previous study[29]. Then the cell lysates and the purified fusion proteins were subjected to 12.5% SDS-PAGE and Western blot analysis.
Immunization of mice
A group of five 7-week-old female Balb/C mice were immunized intraperitoneally with 25 µg of His-preS1 (21-119) fusion proteins[30] in 0.5 mL of complete Freund’s adjuvant (CFA, Sigma) at wk 0 and boosted at wk 2, 4, 6, and 8 with a same amount of antigen in Freund’s incomplete adjuvant (IFA, Sigma). The collected sera obtained at wk 0, 1, 3, 5, 7 and 9 were used to analyze the antigenicity of the above eight preS1 fusion proteins by Western blot analysis and ELISA.
Western blot analysis
After electrophoresis, electrophoretic protein bands of the expressed cell lysates were transferred to nitrocellulose membranes (Hoefer Scientific). The membranes were incubated with the mixture of five mice antisera (1:300 dilution) collected at wk 9, then reacted with goat anti-mouse IgG peroxidase conjugates (Novagen) (1:20000 dilution). After being washed, the substrate solution containing DAB·4HCl (0.5 mg/mL, Amersco) and H2O2 (0.1 µL/mL) was added and the reaction was quenched with distilled water.
ELISA of mouse antisera
A same amount (100 ng/well) of eight preS1 fusion proteins dissolved in 15 mmol/L sodium carbonate, 35 mmol/L sodium bicarbonate buffer, pH 9.6 were coated on microtiter plates (Nunc) at 37 °C for 2 h. After being washed thrice with PBS-0.1% Tween 20 (PBS-T) and blocked with 2.5% bovine serum albumin in PBS at 37 °C for 1 h, separate mouse antisera (1:300 dilution) collected at different time points were added and incubated at 37 °C for 1 h. The wells were washed five times as described above, then goat anti-mouse IgG peroxidase conjugates (Novagen) (1:20000 dilution) were added and incubated at 37 °C for 1 h. The substrate solution (0.06 mg/mL TMB and 0.3 µL/mL H2O2 in 0.1 mol/L Na2HPO4-citrate buffer, pH 5.0) was added to each well and incubated at 37 °C for 15 min. The reaction was stopped with 2 N sulfuric acid and A values were measured at 450 nm in an ELISA reader. The mean A value from five mice antisera was plotted on the corresponding graph.
In competitive ELISA, the mouse antisera were diluted in different concentrations, and previously incub-ated with the coated antigens or other overlapping antigen (s) at a dose of 2 µg at 37 °C for 1 h. The antisera without antigen treatment were used as controls. The selection of coated antigens with immunogenicity depends on the results of Western blot analysis. The other experimental steps were the same as described above.
ELISA of human sera
The plate wells were coated with different preS1 fusion proteins as described above, and then washed thrice with PBS-0.5% Tween 20 (PBS-T) and blocked with 25% goat sera, 2.5% skimmed milk in PBS at 37 °C for 1 h. Human serum samples from 28 normal individuals, 37 acute hepatitis B patients, 106 chronic hepatitis B patients and 33 HCC patients with hepatitis B collected from Changzheng Hospital, Shanghai and East Hepatobiliary Surgery Hospital, Shanghai were diluted 1:30 with sample buffer (25% goat sera, 2.5% skimmed milk, 10% glycerol, 0.19% EDTA in PBS-T). The plates were filled with the diluted serum samples and incubated at 37 °C for 1 h, then washed seven times as described above. Peroxidase conjugated goat anti-human IgG (γ-chain specific, Sigma) (1:5000 dilution) was added to each well and incubated at 37 °C for 1 h. The plates were then washed seven times with PBS-T and the substrate solution was added to react. After 15-min incubation, the reaction was stopped with 2 N sulfuric acid. A values were measured at 450 nm in an ELISA reader. The mean value obtained from the sera of different infection types was plotted on the corresponding graph.
In competitive ELISA, the mixture of serum samples from acute hepatitis B patients was used and the other procedures were similar to those in mice.
RESULTS
Expression, purification of GST-preS1 fusion proteins
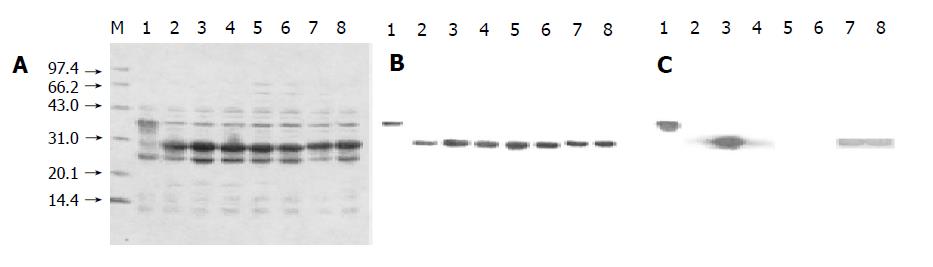
After the expression plasmids were verified by sequencing, they were transformed into E.coli BL21 (gold). The eight overlapping preS1 fusion proteins were highly expressed in the transformed cells (Figure 1A). The supernatants of sonicated cell lysates were applied to affinity chromat-ography on glutathione-sepharose 4B column, and the purified fusion proteins were obtained (Figure 1B). The purities of purified fusion proteins were evaluated to be over 95% by densitometric scanning. About 50-90 mg of soluble purified proteins was obtained from 1 L of culture.
Figure 1 Expression, purification, and Western blot analysis of eight GST fusion proteins containing overlapping preS1 fragments in E.
coli BL21 (gold). A and B: Cell lysates and purified fusion proteins subjected to 12.5% SDS-PAGE and stained with Coomassie brilliant blue. C: Protein bands on the duplicate gel of cell lysates electroblotted onto a nitrocellulose membrane for Western blot analysis with mice antisera. Lane M: Molecular weight standard. Lanes 1-8: The samples containing preS1 (21-119), preS1 (21-47), preS1 (34-59), preS1 (48-70), preS1 (60-85), preS1 (71-94), preS1 (86-109) and preS1 (95-119) in a sequent order.
Immune response to His-preS1 (21-119) fusion protein in mice
The results of Western blot analysis (Figure 1C) showed that the immunogenic domains of preS1 (21-119) sequences in mice existed in the fragments of preS1 (21-47), preS1 (34-59) and preS1 (48-70) in the N-terminus, and in the fragments of preS1 (86-109) and preS1 (94-119) in the C-terminus, whose strips appeared obvious staining intensity. Especially, the staining intensity of preS1 (34-59) strips was much higher than that of the other preS1 fragments. While the staining intensity of preS1 (60-85) and preS1 (71-94) strips was very weak, suggesting that the five fragments in preS1 (21-119) region, especially preS1 (34-59), had strong immunogenicity, and the two fragments in the middle had little immunogenicity.
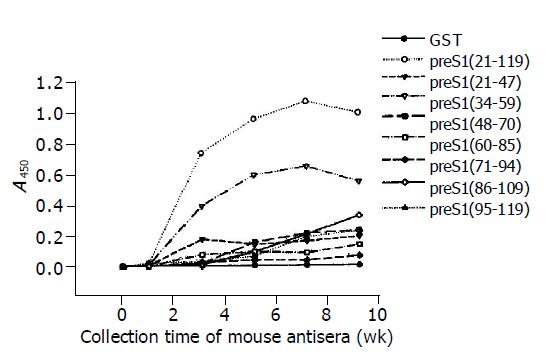
The results of ELISA as shown in Figure 2 indicated that the immunoreactivity of antisera directed against most preS1 fragments increased obviously at wk 3 and went to the utmost at wk 7. It also showed that the antigenicity of preS1 (21-119) and preS1 (34-59) decreased slightly after wk 7, while the antigenicity of the other fragments did not. The distribution and strength of immunogenicity in these fragments were in agreement with the observations in Western blot analysis. The results also demonstrated that preS1 (34-59) fragments were also the strongest in immu-nogenicity.
Figure 2 Immunogenicity analysis of different fragments in preS1 (21-119) region in mice by ELISA using eight overlapping GST-preS1 fusion proteins as coated antigens.
The mice were immunized with His-preS1 (21-119) fusion proteins at wk 0, 2, 4, 6 and 8, and the antisera were collected the following week.
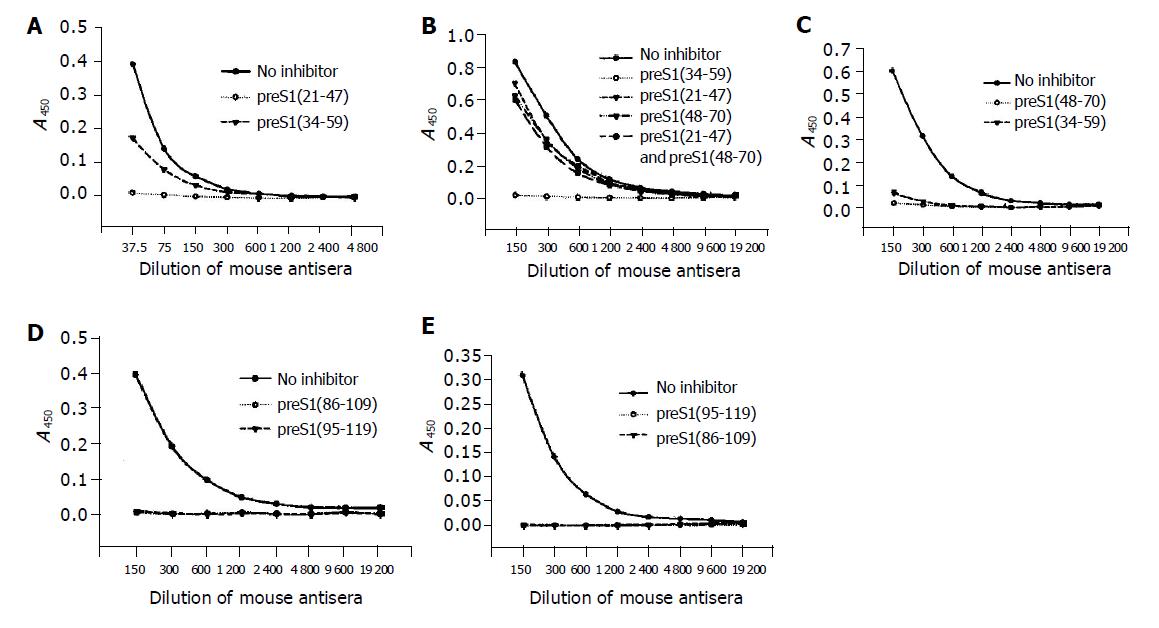
The precise immunogenic domains in preS1 (21-119) region in mice were further identified with competitive ELISA. The antigenicity of each coated antigen could be inhibited by itself, which confirmed the specificity of its corresponding antibody. The antigenicity of preS1 (21-47) could be partially inhibited by preS1 (34-59) (Figure 3A), suggesting that the two fragments, preS1 (21-33) and preS1 (34-47) had immunogenicity. The antigenicity of preS1 (34-59) could only be slightly inhibited by preS1 (21-47) or preS1 (48-70) or a mixture of them (Figure 3B), implying that there might be a conformational epitope in preS1 (34-59) region. While the antigenicity of preS1 (48-70) could be almost totally inhibited by preS1 (34-59) (Figure 3C), suggesting that there was an immunogenic domain in the part of preS1 (48-59). In addition, the antigenicity of preS1 (86-109) and preS1 (95-119) could be inhibited by each other (Figure 3D and 3E), implying that the immunogenic domain existed in the overlapping part, preS1 (95-109).
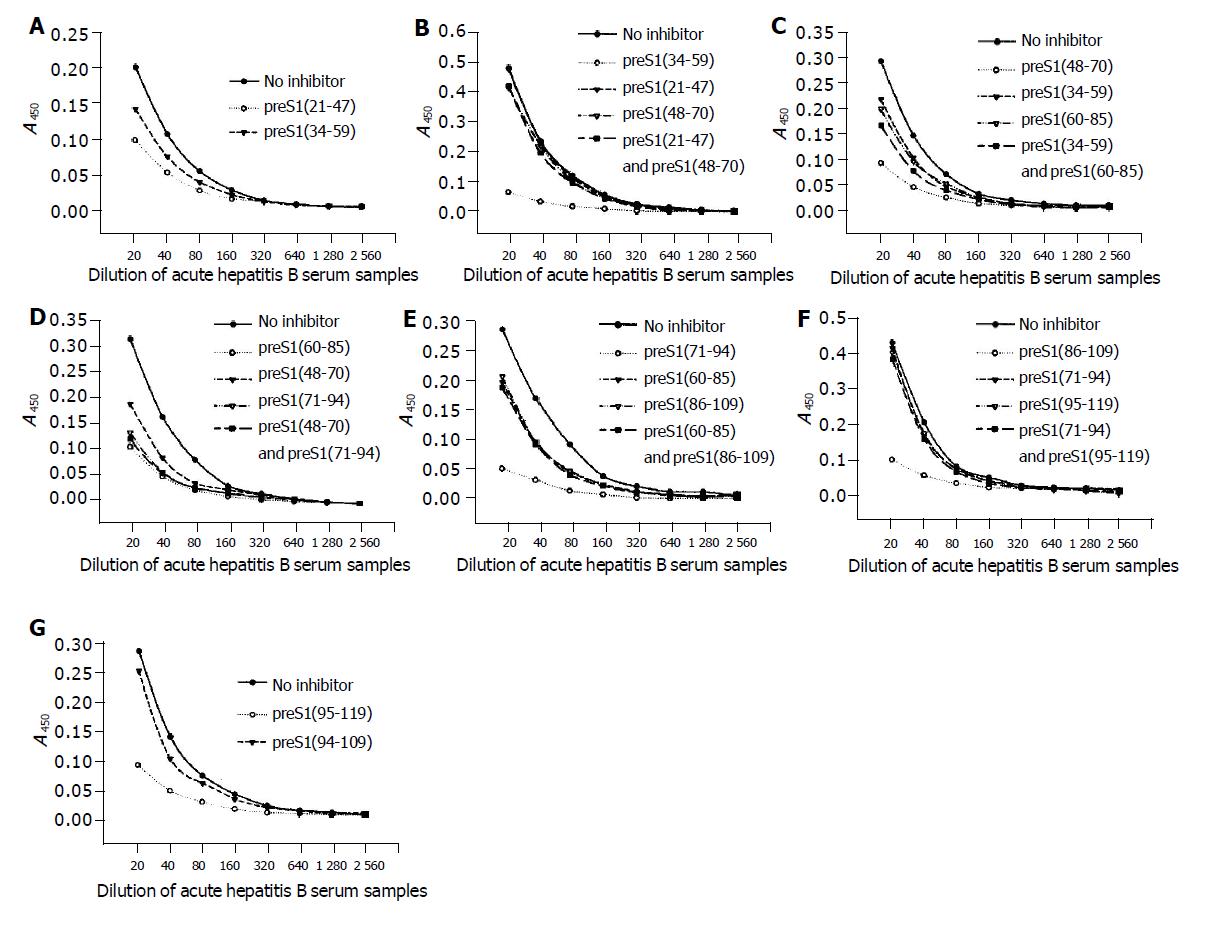
Figure 3 Identification of the precise immunogenic domains in mice by competitive ELISA.
The coated antigens were the five fusion proteins containing different preS1 fragments: preS1 (21-47) (A), preS1 (34-59) (B), preS1 (48-70) (C), preS1 (86-109) (D) and preS1 (95-119) (E), respectively. The mouse antisera colleted at wk 9 were previously incubated with the coated antigens or the other overlapping antigen(s) at a dose of 2 µg, and the samples not treated with antigen(s) were used as controls.
Detection of antibodies against overlapping preS1 fragments in the sera from HBV-infected patients
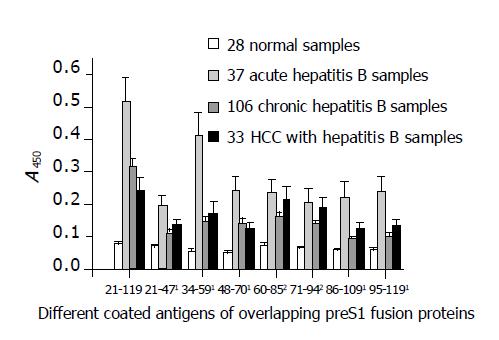
The activity of antibodies against different preS1 fragments detected from the sera of patients with different infection types is shown in Figure 4. The discrimination of HBV infection types was based on the immunological evidence of sera and clinical manifestations of cases. The results indicated that all the fragments showed stronger antigenicity against acute hepatitis B patient samples and the fragments of preS1 (34-59) also had much stronger antigenicity as in mice. In addition, the antigenicity of the fragments, preS1 (21-47), preS1 (48-70), preS1 (86-109), preS1 (95-119), and especially preS1 (34-59), was significantly lower in chronic-phase sera (chronic hepatitis B and HCC with hepatitis B samples) than in acute-phase sera (P<0.05). However, the antigenicity of the fragments, preS1 (60-85) and preS1 (71-94), in the middle part of preS1 showed less diversity (P>0.05).
Figure 4 Detection of antigenicity (A450) of the eight different preS1 fragments overlapped in preS1 (21-119) region against hepatitis B patients’ serum samples with different infection types by indirect ELISA.
1The antigenicity of the fragments, preS1 (21-47), preS1 (48-70), preS1 (86-109), preS1 (95-119), and especially preS1 (34-59), decreased significantly in chronic-phase sera (chronic hepatitis B and HCC with hepatitis B samples) in comparison with acute-phase sera (P<0.05). However, 2the antigenicity of the fragments, preS1 (60-85) and preS1 (71-94), in the middle part showed less diversity (P>0.05). Each column represents the mean A value±SE.
The data of competitive ELISA (Figure 5) likewise confirmed the specificity of corresponding antib-odies. It also indicated that the antigenicity of most fragments could not be chiefly inhibited by the overlapping fragment(s). With an exception, the antigenicity of preS1 (60-85) could be most inhibited by preS1 (71-94), but could not be inhibited by preS1 (48-70). The above results suggested that all the domains except for preS1 (60-70) had some immunogenicity.
Figure 5 Identification of the precise immunogenic domains in humans by competitive ELISA.
The coated antigens were the seven fusion proteins containing different preS1 fragments: preS1 (21-47) (A), preS1 (34-59) (B), preS1 (48-70) (C), preS1 (60-85) (D), preS1 (71-94) (E), preS1 (86-109) (F) and preS1 (95-119) (G), respectively. The mixture of sera samples from acute hepatitis B patients was previously incubated with the coated antigens or the overlapping antigen(s) at a dose of 2 µg, and the samples not treated with antigen(s) were used as controls.
DISCUSSION
Because the HBsAg preS1 region correlates with the assembly and infectivity of HBV[31-33], possesses the binding site on hepatocyte membranes[31], has abundant T-and B-cell epitopes[13] and may overcome the none-response against common vaccines containing only S region[34,35], the incorporation of preS1 region into epitope-based vaccines has been widely accepted[10,36]. While the incorporation site and size of preS1 sequences are controversial, some prev-ious studies on the immunogenicity of preS1 region mainly focused on mice and the tested antigens in preS1 region did not overlap[13,37]. So it may acquire more information about the immunogenic domains in preS1 region if its immu-nogenic domains in mice and humans with overlapping preS1 fragments are simultaneously analyzed.
It has been established that small proteins could be successfully expressed in E.coli by fusing them with GST[38], with which the fusion proteins could be expressed mostly in soluble form and purified more easily by a simple affinity chromatography. In addition, GST did not influence the specificity of anti-preS1 antibodies[29,39]. Therefore, we selected the plasmid of pGEX 4T-1 as a vector to express the different overlapping preS1 fragments in E.coli and gained the satisfactory results. Then the mice were immunized by His-preS1 (21-119) fusion proteins expressed previously in our laboratory[30] and the anti-sera were used to identify the immunogenic domains of preS1 (21-119) region in mice by ELISA using these overlapping preS1 fusion proteins. The antigenicity of these different preS1 fragments against the sera from different HBV-infected patients was also invest-igated.
The results of immunogenicity analysis in mice showed that the immunogenic domains existed in preS1 (21-59) and preS1 (95-109). It is in agreement with the previous observ-ations, which indicated that the immunogenic domains of preS1 in mice were located in preS1 (16-53) and preS1 (94-117) with the immunization of HBsAg/p43 particles[13]. A previous study also found that the immunogenicity of natural and recom-binant antigens was identical[40]. However, the results in humans indicated that other fragments, such as preS1 (71-94), also had strong immunogenicity. It implies that there are more abundant immunogenic domains in preS1 region and more complex immune responses to preS1 region in humans, which might be due to the more complex MHC gene regulation of human immune responses[41]. Additionally, the antigenicity of the N-and C-terminal of the sequences of preS1 (21-119), preS1 (21-47), preS1 (48-70), preS1 (86-109), preS1 (95-119) and especially preS1 (34-59), was significantly lower in chronic-phase sera than in acute-phase sera (P<0.05), leading to the speculation that the antibodies against these fragments, especially preS1 (34-59), are virus-neutralizing.
On the other hand, some previous data figured out that the major epitope of preS1 region in acute-phase sera was conformational, its specific antibody was virus-precipitating and the activity was decreased later[37,40-42]. But it has not been identified. Importantly, the results here clearly showed that the fragments of preS1 (34-59) were the major immu-nogenic domains in mice and human immune responses to preS1 region and its antibody activity was also decreased in mice at wk 7 after immunization. Therefore, the antibody could be speculated to be virus-neutralizing, and it might be efficient to develop a promising vaccine, which incorporates these fragments into HBsAg-based recombinant vaccines.
The predictions of secondary structure and immunological properties in the preS1 (21-119) region using Lasegene software (DNASTAR Inc., Madison, WI) also supported the above results. In conclusion, there are a great deal of turn regions and coil regions, which lack of alpha and beta regions in preS1 region. The only alpha region is located in the domain of preS1 (51-58), which may partially explain the much stronger immunogenicity of preS1 (34-59). In addition, the N-and C-terminal in preS1 (21-119) region have a strong hydrophilicity, a high antigenic index and a high surface probability, which cause the strong immuno-genicity, while preS1 (60-70) has a very weak hydrophilicity, a low antigenic index and low surface probability, which cause the little immunogenicity.