Published online Mar 14, 2005. doi: 10.3748/wjg.v11.i10.1457
Revised: August 31, 2004
Accepted: September 3, 2004
Published online: March 14, 2005
AIM: To identify whether the polymorphisms of the N-acetyltransferase (NAT) genes are susceptible to primary liver cancer (PLC) in Luoyang, a PLC low-incidence area of China.
METHODS: The NAT1 and NAT2 genotypes of 96 PLC cases and 173 controls were determined by PCR-RFLP. Both interaction between NAT1 or NAT2 and environmental risk factors were analyzed based on case control study.
RESULTS: Compared to the control group, the frequencies of alleles NAT1*3, NAT1*4, NAT1*10, NAT1*14B and alleles NAT2*4, NAT2*6, NAT2*7 in PLC group showed no statistically significant difference (χ2 = 2.61 and 4.16, respectively, both P>0.05). The frequencies of NAT1 genotypes NAT1*3/*3, NAT1*3/*4, NAT1*3/*10, NAT1*3/*14B, NAT1*4/*4, NAT1*4/*10, NAT1*4/*14B, NAT1*10/*10, NAT1*10/*14B, and NAT2 genotypes NAT2*4/*4, NAT2*4/*6, NAT2*4/*7, NAT2*6/*6, NAT2*6/*7 and NAT2*7/*7 also had no statistically significant difference between the two groups (χ2 = 11.86 and 2.94 respectively both, P>0.05). Neither the frequencies of rapid and slow NAT1 acetylators nor the frequencies of rapid and slow NAT2 acetylators were significantly different between the two groups (χ2 = 0.598 and 0.44, respectively, both P>0.05). The interaction between NAT1*10 and occupational exposures was found significant with an odds ratio of 3.40 (χ2 = 8.42, P = 0.004, OR 95%CI:1.03-11.22). But no interaction was found between NAT2 and any environmental risk factors.
CONCLUSION: The polymorphisms of NAT1 and NAT2 are not susceptible to PLC in Luoyang. Allele NAT1*10 interacts with occupational exposures.
- Citation: Zhang XF, Bian JC, Zhang XY, Zhang ZM, Jiang F, Wang QM, Wang QJ, Cao YY, Tang BM. Are polymorphisms of N-acetyltransferase genes susceptible to primary liver cancer in Luoyang, China? World J Gastroenterol 2005; 11(10): 1457-1462
- URL: https://www.wjgnet.com/1007-9327/full/v11/i10/1457.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i10.1457
Primary liver cancer (PLC), most of which refers to primary hepatocellular carcinoma (HCC), following lung cancer and stomach cancer, ranked the third of cancer mortality worldwide, accounting for an estimated 357000 deaths in 1990[1]. In China, PLC ranked the second of cancer mortality since 1990s[2]. The incidence of PLC in China has also been found to be increasing, covering about 42.5% of the new global 260000 cases each year[3]. To explore the risk factors of PLC in mainland China has remained an attractive target.
The pathogenesis of PLC remains unknown although much progress has been made in the past decades. Many studies have suggested that PLC is a multi-factorial disease induced by interactions between genetic and environmental factors. Researches in molecular genetics have revealed that polymorphisms of some metabolizing enzyme genes may related to the development of PLC. N-acetyltransferase (NAT) is a kind of enzyme family, which catalyzes acetylation reactions of nitrogenous compounds. It can activate or inactivate extrinsic nitrogenous substances especially amine carcinogens[4]. NAT is encoded by two isozyme genes, NAT1 and NAT2.
Much work has been reported on the relationship between NAT1 genetic polymorphisms and some cancers such as bladder or colon cancer, a few of which were focused on PLC in high incidence areas. In contrast, little has been reported on association between NAT2 genetic polymorphisms and PLC. Bian et al[5] who studied the genotypes, phenotypes and polymorphism of NAT2 for 65 HCC cases in high incidence area of China, found out that slow NAT2 acetylators (NAT2*4 allele) increased the risk of HCC. However, no such works have been reported in PLC low incidence areas of China or other countries. In order to know whether polymorphisms of NAT1 and NAT2 are susceptible to PLC, and to know the interaction between environmental risk factors and NAT1 or NAT2 in such areas, we selected Luoyang City as our research site, which is located in the west of Henan Province, China.
We collected 96 newly diagnosed PLC patients as the case group from more than ten hospitals of Luoyang city during 1999-2002. All the patients met the diagnosis criteria for PLC established by Chinese Anti-Cancer Association[6]. Meanwhile, we collected 65 controls from hospitals and another 108 controls from communities who were all excluded for any hepatic diseases or cancers. Hospital and community controls were merged as the control group based on their balance in gender, age and resident area. All the controls had no kinship with each other. Both groups were of local residents.
All of the subjects gave verbal informed consent for the survey. A same questionnaire was applied to both groups to investigate their general information, health condition, smoking history, occupational exposures etc. We defined smoking as consuming at least one cigarette each day for more than 6 mo, and occupational exposures as contact history with suspicious carcinogens such as benzene, paint and dust.
All cases and controls were interviewed and their medical records were reviewed by trained field workers. They also had 5 mL peripheral blood drawn, which was non-coagulated and kept in -20 °C.
Genomic DNA was extracted from peripheral blood leucocytes by general phenol-chloroform method.
Primers were synthesized based on relevant literatures[7,8] (Table 1). The NAT1 amplification reaction was carried out in a 30 µL solution containing 0.1-0.3 µg DNA, 2.5 mmol/L 4×dNTP (Shanghai Bioasia Company), 0.2 µmol/L NAT1a and NAT1b primer each (Shanghai Sangon Company), 1.5 U Taq DNA polymerase (Beijing Sbsbio Company) and 3.0 µL 10×PCR reaction buffer. The PCR condition consisted of an initial denaturation at 94 °C for 5 min and extension at 72 °C for 7 min followed 30 cycles of denaturation at 94 °C, annealing at 55 °C and extension at 72 °C each for 60 s.
| Primer | Position | Length (bp) | Primer sequence |
| NAT1a | 10-29 | 1158 | 5’-TTAGGAATTCATGGACATTGAAGCATATCTTGAAAGAAT-3’ |
| NAT1b | 1127-1148 | 5’-GCTTTCTAGCATAAATCACCAA -3’ | |
| NAT2a | 1-30 | 895 | 5’-ATGGACATTGAAGCATATTTTGAAAGAATT-3’ |
| NAT2b | 867-895 | 5’-AAGGGTTTATTTTGTTCCTTATTCTAAAT-3’ |
For NAT2, the amplification reaction was carried out in a 100 µL solution containing 1-3 µg DNA, 2.4 mmol/L 4×dNTP, 0.3 µmol/L NAT2a and NAT2b primer each, 0.8 µL Taq DNA polymerase (5 U/µL) and 10 µL 10×PCR reaction buffer. The PCR condition consisted of an initial denaturation at 94 °C and extension at 72 °C each for 5 min followed 30 cycles of denaturation at 94 °C, annealing at 55 °C and extension at 72 °C each for 60 s.
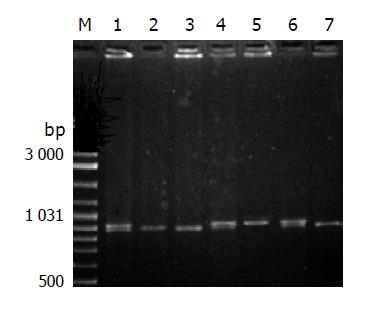
The 5-20 µL of NAT1 PCR product was then digested by Alw26I, AseIand HinfI, and NAT2 by TaqI and Bam HI, for 12-16 h at 37 °C water. The reaction products were separated by electrophoresis under 200 V of constant voltage for 30 min through 2.5-3.0% agarose gel with 10 mg/µL ethidium bromide, which was examined under ultraviolet irradiation.
The genotypes of single nucleotide polymorphisms (SNPs) of NAT1 at nucleotide 560, 1088 and 1095, and SNPs of NAT2 at nucleotide 590 and 857 were determined and divided into wild homozygote (w/w), heterozygote (w/m) and mutant homozygote (m/m) according to the pictures of electrophoresis. Then the genotypes of NAT1 (NAT1*3, *4, *10, *14A, 14B) and NAT2 (NAT2*4, 6, 7) of all subjects were determined based on relevant literature.
The NAT1*10 phenotype and its association with cancer susceptibility remain poorly understood. Commonly NAT1*10 may increase enzyme activity and was found to increase risk of some cancers. In this study individuals with NAT1*10 were classified as rapid NAT1 acetylators and others were slow NAT1 acetylators as usual[9]. As for NAT2, those without NAT2*4 could increase susceptibility to some cancers, and were regarded as slow NAT2 acetylators, those with NAT2*4 were rapid acetylators[8].
Data are presented as mean±SD. Database was built by Epi Info 6.0 and the analysis of data was accomplished using SPSS 10.0 software. The genotype distribution of NAT1 and NAT2 in controls was compared with that as expected from Hardy-Weinberg equilibrium by χ2 tests. The difference in frequency distributions of genotypes and phenotypes between the two groups was tested by χ2 test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to assess the strength of interaction between environmental risk factors and polymorphisms of NAT1 or NAT2. P values of less than 0.05 were considered to be statistically significant.
In the case group, the age span was 20-75 years old, mean age was (58.20±10.71) years old, and gender ratio was 3:1. While in the control group, the age span was 26-80 years, mean age was (57.68±10.03) years old and gender ratio was 2.5:1. Case group and control group had similar frequency for distribution of age and sex (P>0.05). As for the control group, the frequencies of NAT1 and NAT2 genotypes fitted well with the Hardy-Weinberg equilibrium, indicating that the control group were sufficiently representative (Tables 2, 3).
| Genotypes | Observed number | Expected number |
| NAT1*3/ *3 | 20 | 11 |
| NAT1*3/ *4 | 30 | 40 |
| NAT1*3/ *10 | 15 | 22 |
| NAT1*3/ *14B | 4 | 4 |
| NAT1*4/ *4 | 50 | 37 |
| NAT1*4/ *10 | 27 | 39 |
| NAT1*4/ *14B | 5 | 6 |
| NAT1*10/ *10 | 19 | 10 |
| NAT1*10/ *14B | 4 | 3 |
| Total | 173 | 173 |
| Genotypes | Observed number | Expected number |
| NAT2*4/*4 | 100 | 91 |
| NAT2*4/*6 | 23 | 33 |
| NAT2*4/*7 | 28 | 34 |
| NAT2*6/*6 | 6 | 4 |
| NAT2*6/*7 | 12 | 7 |
| NAT2*7/*7 | 4 | 4 |
| Total | 173 | 173 |
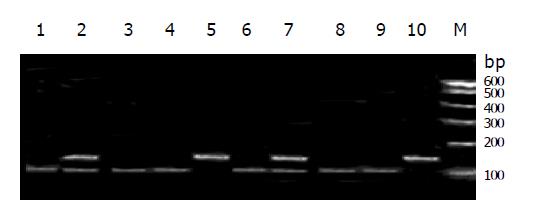
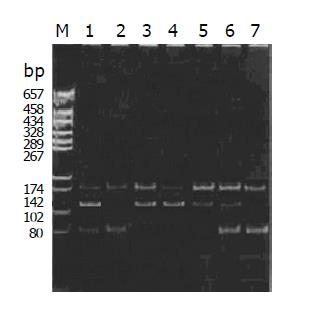
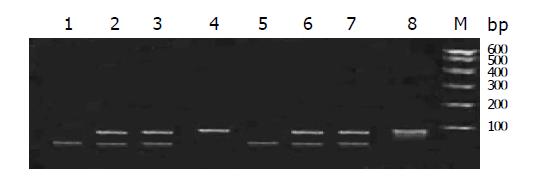
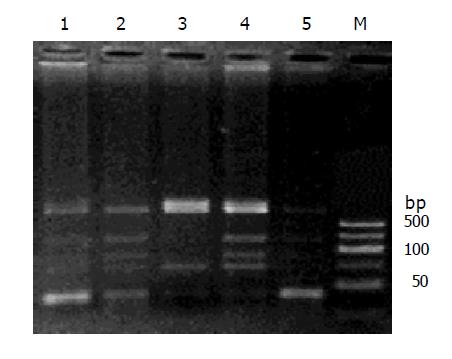
The RFLP results of NAT1 and NAT2 Figures 1, 2, 3, 4, and 5. Neither 3 SNPs of NAT1 (C1095A, T1088A and G560A) nor 2 SNPs of NAT2 (G857A and G590A) were found to be associated to PLC (χ2= 1.170, 0.190, 0.047, 3.33 and 0.95, respectively, all P>0.05).
Similarly, we did not find any association between PLC and NAT1 or NAT2 alleles (χ2 = 2.61 and 4.16, both P>0.05). We failed to observe NAT1*14A alleles in both groups. The genotypes of NAT1 and NAT2 also showed no association with PLC (Tables 4, 5).
| Genotypes | Cases (%) | Controls (%) | Total |
| NAT1*3/ *3 | 4 (4.2) | 20 (11.5) | 24 |
| NAT1*3/ *4 | 24 (25.0) | 30 (17.2) | 54 |
| NAT1*3/ *10 | 3 (3.1) | 15 (8.6) | 18 |
| NAT1*3/ *14B | 4 (4.2) | 4 (2.3) | 8 |
| NAT1*4/ *4 | 30 (31.3) | 50 (28.7) | 80 |
| NAT1*4/ *10 | 10 (10.4) | 27 (15.5) | 37 |
| NAT1*4/ *14B | 3 (3.1) | 5 (2.9) | 8 |
| NAT1*10/ *10 | 14 (14.7) | 19 (10.9) | 33 |
| NAT1*10/ *14B | 4 (4.2) | 4 (2.3) | 8 |
| Total | 96 (100.0) | 173 (100.0) | 269 |
| Genotypes | Cases (%) | Controls (%) | Total |
| NAT2*4/*4 | 57 (59.4) | 100 (57.8) | 157 |
| NAT2*4/*6 | 9 (9.4) | 23 (13.3) | 32 |
| NAT2*4/*7 | 15 (15.6) | 28 (16.2) | 43 |
| NAT2*6/*6 | 2 (2.1) | 6 (3.5) | 8 |
| NAT2*6/*7 | 8 (8.3) | 12 (6.9) | 20 |
| NAT2*7/*7 | 5 (5.2) | 4 (2.3) | 9 |
| Total | 96 (100.0) | 173 (100.0) | 269 |
The phenotypes of NAT1 and NAT2 also showed no difference in frequencies between case group and control group (Table 6), suggesting that the phenotypes of NAT1 and NAT2 were not associated with PLC.
The metabolism of carcinogenic arylamines in tobacco smoke, occupational exposure, fried food or meat is mediated by enzymes including NAT1 and NAT2. Both NAT1 and NAT2 are genotypically and phenotypically polymorphic with variable genotype frequencies in different ethnic groups.
In our case control study design, we classified all the environmental risk factors into two levels (0 and 1); then calculated odds ratios (OR) of different levels among both groups after setting down those without NAT1*10 and without exposures as referential baseline. We then judged the interaction between susceptible allele and environmental carcinogens. The results suggested that interaction between NAT1*10 and occupational exposures had statistical significance with an OR of 3.40 (Table 7). Interactions between NAT1*10 and other risk factors showed no statistical significance. We also analyzed relationship between HBV infection and NAT1*10, but failed to get positive result.
| Occupationalexposures | NAT1*10 | Cases | Controls | OR | 95%CI |
| 51 | 124 | 1 | |||
| + | 11 | 34 | 0.79 | 0.37-1.67 | |
| + | 15 | 15 | 2.43 | 1.11-5.34 | |
| + | + | 7 | 5 | 3.4 | 1.03-11.22 |
As for NAT2, we analyzed interaction between NAT2 and environmental risk factors by case-only study. That is, in the case group, both genotypes and environmental risk factors were classified into two levels (0 and 1), then OR and its 95% CI were calculated. However, we failed to find any positive results. In addition, we applied case control study design method as the above for NAT1 to analyze the interaction between HBV infection and NAT1 or NAT2 genotypes. We found that there was no relationship between NAT1 genotypes and HBV infection. After being stratified by HBV infection, NAT2 genotypes were found to be related to PLC, which suggested that HBV infection interacted with NAT2 genotypes (Table 8). But it needed to be verified because the sample in each level was very small.
| Genotypes | HBV infection (-) | HBV infection (+) | ||
| Cases | Controls | Cases | Controls | |
| NAT2*4/*4 | 33 | 100 | 24 | 1 |
| NAT2*4/*6 | 5 | 24 | 4 | 2 |
| NAT2*4/*7 | 6 | 28 | 11 | 0 |
| NAT2*6/*6 | 1 | 5 | 1 | 1 |
| NAT2*7/*7 | 6 | 4 | 1 | 0 |
| NAT2*6/*7 | 2 | 8 | 2 | 0 |
As an important enzyme involved in the phaseII biotransformation, NAT metabolically activates or inactivates aromatic amines through acetylation. NAT has two isozymes, NAT1 and NAT2. Both genes are located on human chromosome 8p21.3 and 8p23.1 respectively, and their coding area owns 87% homology.
To select a low incidence area of PLC in mainland China as research field to analyze the relationship between NAT1 and NAT2 genetic polymorphisms and PLC is a new attempt. We should explain that some of our cases had accepted chemotherapy or radiotherapy before our research. Considering that genes are stable and not so changeable genetic substances, and reports showed that the null GSTM1 genotype frequency of HCC patients were not affected by chemotherapy or radiotherapy[10], we did not take into account the effect of these two therapies on NAT genotypes. In this study, the genotype frequencies of NAT1 and NAT2 in the control group fit the Hardy-Weinberg equilibrium very well, suggesting that the study sample was good enough to represent the source communities of Luoyang city.
Compared with relevant research in Taiwan[11], the frequencies of NAT1*3 and NAT2*4 alleles are higher, while frequency of NAT1*10 is lower in both groups than those of Taiwanese. In the control group, the frequency of NAT2*6 is lower than those of Taiwanese. Other alleles are commonly the same between the two ethnic groups. This kind of similarity and difference suggest that NAT genetic polymorphisms could be variable in the same race but of different geographic areas.
Compared to other Chinese people, NAT1*3, NAT1*4 and NAT1*10 allele frequencies of Luoyang people showed little difference, while NAT2*4 and NAT2*6 showed much difference from them[12,13]. Compared to other ethnic groups such as the whites, blacks or Japanese, the NAT1 and NAT2 alleles of Luoyang people are quite different[14-16]. This means that different race or ethnic group owns different genetic composition.
NAT1*10 has been verified to be associated with many cancers and regarded as a high activity allele, whose phenotype is rapid NAT1 acetylator. While few reports could be found on PLC and NAT1 genetic polymorphism, Yu et al[17] in Taiwan found that NAT1*10 is not associated with PLC. Our result supports the same argument. Besides, we found that those with NAT1*10 together with occupational exposure has an increased risk of PLC, and the OR is 3.40 (95%CI = 1.03-11.22). Although we failed to find literatures to support this finding, many reports have similar results when analyzing interactions between NAT1*10 and some risk factors in different cancers. For example, a six-fold of increased risk in colorectal adenoma was found among rapid NAT1 acetylators (with NAT1*10 allele) who consume high temperature cooking of red meats often and the OR is 6.50 (95%CI = 2.16-19.7)[18,19]. The association of the NAT1*10 allele with breast cancer was mainly confirmed to former smokers (OR = 3.3, 95%CI = 1.2-9.5). All these suggest that NAT1 affect carcinogenic process through some way.
In this study, we find that NAT2 genetic polymorphisms are not susceptible to PLC. There are some arguments on their relationship however. Bian et al[5] found that in PLC high incidence area of China, slow NAT2 acetylators had an increased risk of PLC. Yu et al[17] investigated all the HBV carriers in HCC cases and controls, finding that among smokers, for those with NAT2*4/*4 genotype, the odds ratio of developing HCC was 2.58 (95%CI, 1.04-6.43), for those with NAT2*4 allele, the OR was 2.67 (95%CI, 1.15-6.22), so they concluded that rapid NAT2 acetylators had an increased risk of HCC. Huang et al[20] in Taiwan did not find association between the susceptibility of HCC and the overall NAT2 genotypes. However, the interaction between red meat intake and the NAT2*4 acetylator status for an increased risk of HCC was significant in those with chronic viral hepatitis-related cirrhosis. While Spanish researchers found that slow NAT2 acetylators were susceptible to HCC[21], in Fujian Province of China, the smokers with slow acetylation genotype of NAT2 were found to be the population with high risk for HCC[22]. These two conclusions are similar to that of Bian et al[5]. In our study, slow acetylation genotype of NAT2 was not found to be a risk factor of PLC in Luoyang city, NAT2 genotypes did not interact with environmental risk factors. All of the above results suggest that NAT2 gene may play different roles in hepatocarcinogenesis between PLC high and low incidence areas, mainland China and Taiwan, or among different ethnic groups.
All of the different or similar findings lead to such interpretations: First, NAT1 and NAT2 genetic polymorphisms in low incidence area of PLC do have no association with PLC. Second, different regional or ethnic groups have different exposures to the environment, and the metabolism of carcinogens also involves a number of other enzymes, so it is impossible to explain the hepatocarcinogenesis by only genetic polymorphism of one enzyme and its interaction with environmental risk factors. Third, the sample of our study is not big enough to reveal the real truth of the relationship between NAT polymorphism and PLC.
Although most of our results were negative, our research gained the argument from a whole new perspective that NAT1 and NAT2 genes are not negligible in hepatocarcinogenesis study in different areas. Our study also revealed some new questions that need further worthwhile examination.
Edited by Li WZ Language Editor Elsevier HK
| 1. | Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997;349:1269-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2466] [Cited by in RCA: 2352] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 2. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] |
| 3. | Xiao KY, Peng MH. Advances in epidemiologic study of primary liver cancer. Zhongguo Puwai Jichu Yu Linchuang Zazhi. 2000;7:272-274. |
| 4. | Hein DW. N-Acetyltransferase genetics and their role in predisposition to aromatic and heterocyclic amine-induced carcinogenesis. Toxicol Lett. 2000;112-113:349-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Bian JC, Shen FM, Wang JB, Wu Y, Zhang BC. Relationship between genetic polymorphism of N-acetyltransferase and genetic susceptibility to primary hepatocellular carcinoma. Zhonghua Yixue Yichuanxue Zazhi. 1997;14:16-20. |
| 6. | Chinese Anti-Cancer Association. Diagnostic criteria for primary liver cancer. Zhonghua Ganzangbing Zazhi. 2000;8:135. |
| 7. | Deitz AC, Doll MA, Hein DW. A restriction fragment length polymorphism assay that differentiates human N-acetyltransferase-1 (NAT1) alleles. Anal Biochem. 1997;253:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Doll MA, Fretland AJ, Deitz AC, Hein DW. Determination of human NAT2 acetylator genotype by restriction fragment-length polymorphism and allele-specific amplification. Anal Biochem. 1995;231:413-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Hein DW, Leff MA, Ishibe N, Sinha R, Frazier HA, Doll MA, Xiao GH, Weinrich MC, Caporaso NE. Association of prostate cancer with rapid N-acetyltransferase 1 (NAT1*10) in combination with slow N-acetyltransferase 2 acetylator genotypes in a pilot case-control study. Environ Mol Mutagen. 2002;40:161-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Bian JC, Wang JB, Wu Y, Zhang BC, Shen FM. Relationship between GSTM1 null genotype and genetic susceptibility to pri-mary hepatocellular carcinoma. Zhonghua Yixue Yichuanxue Zazhi. 1996;13:353-356. |
| 11. | Hsieh FI, Pu YS, Chern HD, Hsu LI, Chiou HY, Chen CJ. Genetic polymorphisms of N-acetyltransferase 1 and 2 and risk of cigarette smoking-related bladder cancer. Br J Cancer. 1999;81:537-541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Zhao B, Lee EJ, Yeoh PN, Gong NH. Detection of mutations and polymorphism of N-acetyltransferase 1 gene in Indian, Malay and Chinese populations. Pharmacogenetics. 1998;8:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Lee EJ, Zhao B, Seow-Choen F. Relationship between polymorphism of N-acetyltransferase gene and susceptibility to colorectal carcinoma in a Chinese population. Pharmacogenetics. 1998;8:513-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Millikan RC, Pittman GS, Newman B, Tse CK, Selmin O, Rockhill B, Savitz D, Moorman PG, Bell DA. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7:371-378. [PubMed] |
| 15. | Bouchardy C, Mitrunen K, Wikman H, Husgafvel-Pursiainen K, Dayer P, Benhamou S, Hirvonen A. N-acetyltransferase NAT1 and NAT2 genotypes and lung cancer risk. Pharmacogenetics. 1998;8:291-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 54] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Katoh T, Kaneko S, Boissy R, Watson M, Ikemura K, Bell DA. A pilot study testing the association between N-acetyltransferases 1 and 2 and risk of oral squamous cell carcinoma in Japanese people. Carcinogenesis. 1998;19:1803-1807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Yu MW, Pai CI, Yang SY, Hsiao TJ, Chang HC, Lin SM, Liaw YF, Chen PJ, Chen CJ. Role of N-acetyltransferase polymorphisms in hepatitis B related hepatocellular carcinoma: impact of smoking on risk. Gut. 2000;47:703-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Ishibe N, Sinha R, Hein DW, Kulldorff M, Strickland P, Fretland AJ, Chow WH, Kadlubar FF, Lang NP, Rothman N. Genetic polymorphisms in heterocyclic amine metabolism and risk of colorectal adenomas. Pharmacogenetics. 2002;12:145-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Zheng W, Deitz AC, Campbell DR, Wen WQ, Cerhan JR, Sellers TA, Folsom AR, Hein DW. N-acetyltransferase 1 genetic polymorphism, cigarette smoking, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8:233-239. [PubMed] |
| 20. | Huang YS, Chern HD, Wu JC, Chao Y, Huang YH, Chang FY, Lee SD. Polymorphism of the N-acetyltransferase 2 gene, red meat intake, and the susceptibility of hepatocellular carcinoma. Am J Gastroenterol. 2003;98:1417-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Agúndez JA, Olivera M, Ladero JM, Rodriguez-Lescure A, Ledesma MC, Diaz-Rubio M, Meyer UA, Benítez J. Increased risk for hepatocellular carcinoma in NAT2-slow acetylators and CYP2D6-rapid metabolizers. Pharmacogenetics. 1996;6:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Gao JP, Huang YD, Lin JA, Zhu QC, Liang JP. Relationship between genetic polymorphisms of N-acetyltransferase and susceptibility to hepatocellular carcinoma. Zhonghua GanZangBing ZaZhi. 2003;11:20-22. [PubMed] |