Published online Mar 15, 2004. doi: 10.3748/wjg.v10.i6.841
Revised: October 1, 2003
Accepted: October 7, 2003
Published online: March 15, 2004
AIM: To establish a mice model of hepatitis B by using HBV-transgenic mice, and to transfer HBV-specific cytotoxic T lymphocytes (CTL) induced from syngeneic BALB/c mice immunized by a eukaryotic expression vector containing HBV complete genome DNA.
METHODS: HBV DNA was obtained from digested pBR322-2HBV and ligated with the vector pcDNA3. Recombinant pcDNA3-HBV was identified by restriction endonuclease assay and transfected into human hepatoma cell line HepG2 with lipofectin. ELISA was used to detect the expression of HBsAg in culture supernatant, and RT-PCR to determine the existence of HBV PreS1 mRNA. BALB/c mice were immunized with pcDNA3-HBV or pcDNA3 by intramuscular injection. ELISA was used to detect the expression of HBsAb in serum. MTT assay was used to measure non-specific or specific proliferation ability and specific killing activity of spleen lymphocytes. Lymphocytes from immunized mice were transferred into HBV-transgenic mice (2.5 × 107 per mouse). Forty-eight hours later, the level of serum protein and transaminase was detected with biochemical method, liver and kidney were sectioned and stained by HE to observe the pathological changes.
RESULTS: By enzyme digestion with Eco RI, Xho I and Hind III, the recombinant pcDNA3-HBV was verified to contain a single copy of HBV genome, which was inserted in the positive direction. HepG2 cells transfected with the recombinant could stably express PreS1 mRNA and HBsAg. After immunized by pcDNA3-HBV for 4 weeks, HBsAb was detected in the serum of BALB/c mice. The potential of spleen lymphocytes for both non-specific and specific proliferation and the specific killing activity against target cells were enhanced. The transgenic mice in model group had no significant changes in the level of serum protein but had an obvious increase of ALT and AST. The liver had obvious pathological changes, while the kidney had no evident damage.
CONCLUSION: A eukaryotic expression vector pcDNA3-HBV containing HBV complete genome is constructed successfully. HepG2 cells transfected with the recombinant can express PreS1 mRNA and HBsAg stably. Specific cellular immune response can be induced in mice immunized by pcDNA3-HBV. A mice model of acute hepatitis with HBV has been established.
- Citation: Gao LF, Sun WS, Ma CH, Liu SX, Wang XY, Zhang LN, Cao YL, Zhu FL, Liu YG. Establishment of mice model with human viral hepatitis B. World J Gastroenterol 2004; 10(6): 841-846
- URL: https://www.wjgnet.com/1007-9327/full/v10/i6/841.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i6.841
HBV infection is a worldwide problem that affects human health severely[1-3]. No appropriate animal model is available till now because of the strict host specificity of HBV infection. At present, HBV transgenic animals could be obained during embryonic period, which always induce immune tolerance to dominating HBV antigens[4-6]. In addition, it has great difference in biological characteristics between duck hepatitis B virus and HBV. So it is imperative to establish a mice model that can simulate human viral hepatitis B precisely and is convenient for wider applications.
Both clinical researches and animal experiments suggest that the pathological changes are not caused by the virus directly, but are meditated by immunity[7,8]. HBV specific CTL induced by HBV antigens can cause immune damage to the liver to clear the viruses and recruit non-specific inflammatory cells to local infection tissue simultaneously, thus aggravating hepatic injury[9,10]. We planned to induce HBV specific CTL by gene immunity and to establish a mice model of hepatitis B by transferring CTLs to HBV-transgenic mice.
SPF BALB/c mice were provided by the Laboratory Animal Center of Shandong University. HBV complete genome (ayw subtype) transgenic BALB/c mice with a high level of HBsAg, HBeAg and with HBV DNA positive in peripheral blood, were purchased from Transgenic Animal Central Laboratory, 458 Hospital of PLA, China.
Hepatocellular carcinoma cell line HepG2 was cultured and conserved in our laboratory. HepG2.2.15 cell line carrying four whole HBV genes can not only express HBsAg and HBeAg but also produce Dane’s particles of HBV. SP2/0-HBV is a mouse myeloma cell line with pcDNA3-HBV stably transfected.
pBR322-2HBV was a recombinant plasmid with a double copy of HBV (ayw subtype) inserted into EcoRI restriction site of pBR322. Mammal animal cell expression vector pcDNA3 was 5.4 kb in length. It has a CMV early promoter enhancer, multiple cloning sites (MCS) and neomycin resistant gene.
Alkaline lysis method was used to extract large quantity of pBR322-2HBV and pcDNA3 DNA respectively. Precipitation with polyethylene glycol was used to purify plasmid DNA, and biometer was used for quantitative analysis. pBR322-2HBV was digested with EcoRI and separated by 10 g/L agarose gel electrophoresis. The gel band containing 3.2 kb DNA was cut under UV light. HBV DNA was extracted using a DNA recovery kit. pcDNA3 was also digested by EcoRI and a 5.4 kb fragment was recovered with the same method. Ten microliter of HBV DNA and 2 μL of pcDNA3-HBV (mol ratio 3:1) were mixed together with 2 μL of T4 DNA ligase, 2 μL of buffer and 2 μL of sterile water. The mixture was placed overnight at 4 °C, and then the reaction was terminated. Ten microliter of ligated product was transformed to competent DH5α, which was then transferred to a LB medium plate containing 60 μg/mL of ampicillin and incubated overnight at 37 °C.
Antibiotic selection and restriction endonuclease assay were used to screen and identify positive clones. A positive clone was selected randomly and amplified for recombinant plasmid DNA. Then the recombinant DNA was digested by EcoRI, XhoI and HindIII, respectively. The digestion system was 20 μL, including 1 μL DNA, 1 μL endonuclease, 2 μL10 × buffer and 16 μL sterile water, and incubated for 3 hours at 37 °C. The correctness of ligation was further identified by 10 g/L agarose gel electrophoresis.
HepG2 cells during logarithmic period were digested by trypsinization and diluted to 4 × 105/mL. Then the cells were plated to 24-well plates with 0.5 mL per well (cell count 2 × 105) and cultured for 24 h for transfection next day, when 80-90% of the cells were fused. One μg purified pcDNA3-HBV plasmid DNA was transfected to the prepared HepG2 cells using lipofectin reagent. After transfection, HepG2 cells were screened by G418 resistance and routinely cultured. The culture supernatant was collected periodically to detect HBV antigens. The selected positive clone was named as HepG2.02G.
Concentration of HBsAg in culture supernatant of transfected cells was detected by ELISA. Critical value (cutoff) = 2.1 × N, in which N is the average OD of negative control, and S is the OD of sample. The sample was positive when S/N ≥ 2.1, otherwise negative.
HepG2.02G, HepG2.2.15, and HepG2 cells in logarithmic period were harvested, each of 1 × 106 cells, was rinsed by 0.01 mol/L PBS once and centrifuged at 1 000 rpm for 5 min. The precipitate was used for total RNA extraction according to the protocol of TRIZOL kit.
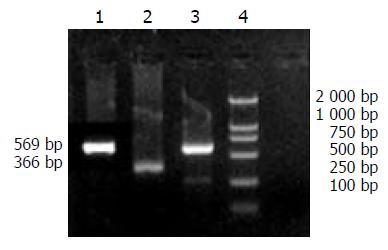
According to the sequence of HBV S1 (subtype ayw) described by GenBank, primers were designed to amplify HBV PreS1 gene. PCR product would be 625 bp. β-actin served as control, with a PCR product 366 bp in length. The sequences of the primers were: PreS1 forward: 5’-GCTCTAGACACCAAATGCCCCTA-3’, reverse: 5’-GCTCTAGAGAATCCCAGAGG-3’, β-actin forward: 5’-ACACTGTGCCCATCTACGAGGGC-3’, reverse: 5’-ATGATGGAGTTGAAGGTAGTT TCG TGG AT-3’. These oligonucleotide primers were synthesized by Shanghai Sangon Biological Engineering Technology and Service Co. Ltd.
Reverse transcription was performed with 1 μg of total RNA to obtain cDNA as the template. PCR was carried out in 25 μL volume with 1 μL RT-PCR product, 2.5 μL 10 × PCR buffer, 0.3 μL Mg2+, 5 nmol/L of dNTP, 3 U Taq polymerase and 10 nmol/L of primers (forward and reverse). The reaction conditions were pre-denatured at 94 °C for 3 min, 30 cycles (denatured at 94 °C for 1 min, annealed at 50 °C for 1 min and extended at 72 °C for 1 min), followed by a final extension at 72 °C for 5 min. Ten microliter of the PCR product then underwent 2% agarose gel electrophoresis at 70 V for 30 min. HepG2 cells were used as negative control, while HepG2.2.15 as positive.
The purified pcDNA3-HBV DNA was diluted with sterile NS to the concentration of 1 000 μg/mL and preserved at -20 °C. Twelve male BALB/c mice were randomly divided into 2 groups, 6 mice each. One was experimental group (Exp), which was injected 50 μL 250 g/L sucrose intramuscularly to both sides of anterior tibial muscle, 50 μL pcDNA3-HBV (100 μL per mouse) was injected at the same site 20 min later. Another was the control group (Ctrl), the same quantity of pcDNA3 was injected as negative control. The immunization was intensified once 2 to 3 wk.
Blood samples were collected by cutting tails of the mice at wk 2, 4, 8 and 10, respectively. Serum was separated and HBsAb was detected by double-antibody sandwich ELISA.
Mice spleen cells were prepared routinely (cell density adjusted to 5 × 106/mL) and plated to 96-well plates (100 μL per well). The supernatant of HepG2.2.15 (containing a high level of HBsAg), PHA (eventual concentration 25 μg/mL) or ConA (eventual concentration 10 μg/mL) was added to 3 wells, respectively. MTT assay was performed after the cells were cultured at 37 °C for 68 h. Proliferation rate was calculated as: (OD of stimulatory – OD of non-stimulatory) / OD of non-stimulatory × 100%.
Mice spleen cells were plated to 96-well plates, mixed with SP2/0-HBV cells (effector/target ratio 50:1). Control groups were designed and each group had 3 wells. MTT assay was performed after the cells were cultured for 72 h. Killing rate of CTLs was calculated as: [1- (OD of effector and target cells - OD of effector cells) / OD of target cells] × 100%.
Transgenic mice were divided into 3 groups (6 mice each): Model, negative control and blank control. Mice in model or negative control group were injected with spleen lymphocytes of mice immunized by pcDNA3-HBV (HBsAb positive) or pcDNA3 respectively. Mice in blank control group were injected with NS, 0.5 mL each mouse. Spleen lymphocytes were adjusted to 5.0 × 107/mL using NS and injected to transgenic mice through caudal vein (2.5 × 107 lymphocytes, 0.5 mL per mouse).
After 48 h, the transgenic mice were killed and their venous blood was collected. Serum total protein, albumin, globulin, ALT and AST were detected. Liver and kidney were fixed with 100 g/L formaldehyde, embedded, sliced and stained with HE to observe inflammation and immune-pathological changes with a microscope.
All data were shown as mean ± SD. Significant levels of differences between values were analyzed using the Chi-square test and Student’s t test. Statistically significant P value was defined as < 0.05.
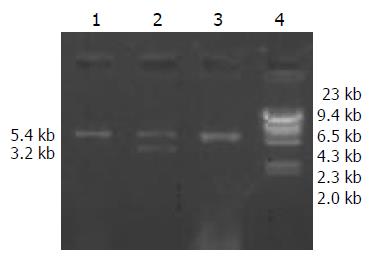
Extracted plasmid DNA had a high purity (OD260/280 = 1.6 - 1.8). After pBR322-2HBV was digested by EcoRI, 2 bands were obtained at about 4.3 kb and 3.2 kb in size (Figure 1). pcDNA3 was linear after digestion, and a band at 5.4 kb proved it was completely digested (Figure 2).
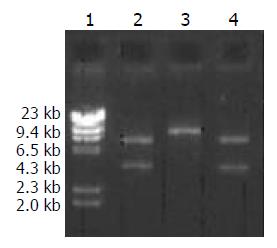
A high purity of the extracted 2 kinds of DNA was certified by electrophoresis. The recombinant pcDNA3-HBV was digested by EcoRI and the product underwent electrophoresis. Only a band of 5.4 kb indicated that the transformed plasmid was pcDNA3 without exogenous genes, while 2 bands at 5.4 kb and 3.2 kb indicated successful ligation (Figure 3). pcDNA3-HBV was digested by HindIII, and the product of 8.6 kb in size indicated that the recombinant pcDNA3-HBV contained a single HBV gene. pcDNA3-HBV was digested by XhoI, and the products of 3.1 kb and 5.5 kb showed that HBV was inserted at the correct direction (Figure 4).
HBsAg in supernatant could be detected on day 15, and expressed stably after 40 d (S/N = 48).
A specific band at 625 bp demonstrated that HBV could efficiently replicate and transcript in human hepatocellular carcinoma cells, as shown in Figure 5.
pcDNA3-HBV immunized mice had specific immune response at the 4th week (S/N = 19). Antibody expression reached the highest level at the 8th week and this level lasted to the 10th week.
As illustrated in Table 1, the non-specific proliferation ability of spleen lymphocytes of mice immunized by pcDNA3-HBV to both PHA and ConA was enhanced as compared with that of pcDNA3 (P < 0.05), and the specific proliferation ability to supernatant of HepG2.2.15 was also promoted (P < 0.01).
MTT assay showed that the killing activity of spleen lymphocytes of mice immunized by pcDNA3 against SP2/0 and SP2/0-HBV had no significant difference (P > 0.05), but that of mice immunized by pcDNA3-HBV to SP2/0-HBV was distinctly enhanced as compared to SP2/0 (P < 0.01). Results are shown in Table 2.
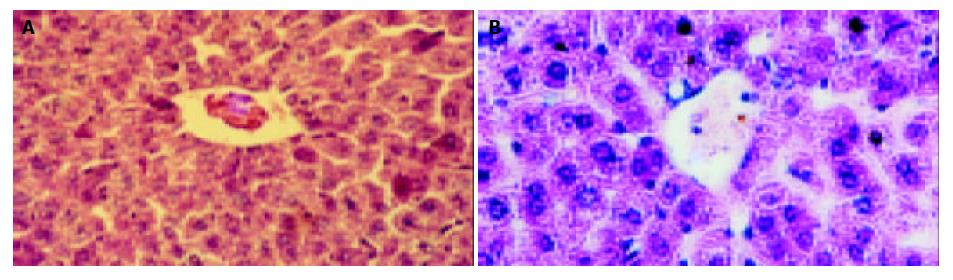
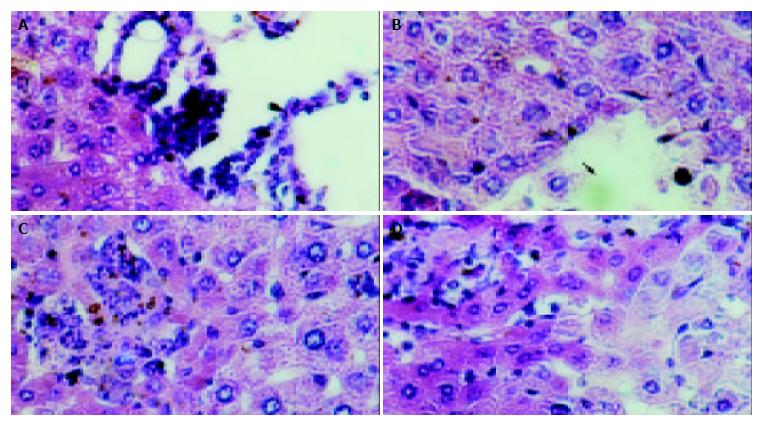
The mice in each group had no significant changes in the level of serum protein compared with the blank control group (P > 0.05). The serum ALT and AST of transgenic mice in blank and negative control groups were normal (0-40 U/L), while they (ALT 232 ± 14 U/L, AST 315 ± 21 U/L) were much higher in the model group than in the control group (P < 0.01). The hepatic cords of mice in the blank control group had a radiated structure surrounding the central vein, the hepatic lobule had a clear structure with cells tightly connected. The hepatic cells had large and round nuclei, with obvious nucleoli and abundant symmetrical cytoplasm. There was no inflammatory cell infiltration in the portal area with a clear structure (Figure 6A). Mice in negative control group had sporadical acidophilic changes of hepatic cells, without inflammatory cell infiltration (Figure 6B). In the model group, hepatic cells of transgenic mice had diffuse ballooning degeneration, cytoplasmic loosening, acidophilic changes, appearance of acidophilic body, spotty necrosis of hepatic tissue and obvious infiltration of inflammatory cells in the portal area (Figure 7). However, mice in the model group had no obvious inflammation and structure damage of the kidney.
The host variety of HBV infection is limited. At present, the animal models for HBV research included chimpanzee[11], Tupaia Glis[12] and other hepadnaviridae virus infected animals[13]. Although duck hepatitis B virus (DHBV) has been found to be similar to HBV in mechanism of pathogenesis, ducks infected with DHBV did not develop hepatic cirrhosis and hepatocellular carcinoma because DHBV lacks X gene[14]. Mice had low susceptibility to HBV, but HBV could replicate in transgenic mice[15]. Two research groups established the transgenic mice which could produce HBV envelopes or HBsAg particles in 1985[16,17]. A high level of HBsAg in circulatory blood could be detected, but no hepatitis was observed in the transgenic mice. The mechanisms of HBV pathogenesis was not caused by virus directly, but closely related to T lymphocyte activation, as demonstrated both in vitro and in vivo[18,19]. Ando et al[20] induced a fulminant hepatitis mice model by transferring HBsAg-specific CTLs to HBsAg-transgenic mice, in which there was no human hepatitis B virus. To establish a mice model of human viral hepatitis B by using HBV complete genome transgenic mice with virus replication in vivo, we constructed a new eukaryotic expression vector containing HBV complete genome and produced HBV-specific CTLs for adoptive transfer by immunizing non-transgenic syngeneic mice with the recombinant.
HBV complete genome DNA was obtained from EcoRI digested pBR322-2HBV and ligated with the vector pcDNA3 which was also digested by EcoRI, thus constructing the recombinant pcDNA3-HBV. pcDNA3 is a eukaryotic expression vector. MCS contains the restriction sites of EcoRI, XhoI and HindIII. CMV promoter can express efficiently in many kinds of cells. The immunostimulatory sequences (ISS) of pcDNA3 can not only activate B lymphocytes to produce antibodies, but also stimulate lymphocytes to secrete IL-12, IFN-γ and IL-6. IL-12 and IFN-γ could foster Th1 responses and enhance cell-mediated immunity, while IL-6 could further promote B lymphocyte activation and antibody secretion and also enhance cell-mediated immunity as its primary role[21,22]. So this vector is a better alternative for gene immunity. The recombinant was constructed by inserting the HBV complete genome to EcoRI sites of pcDNA3, with HindIII site upstream of EcoRI in MCS of pcDNA3 and no HindIII site in HBV genes. A single XhoI site was at 180 bp upstream of HBV gene. According to these characteristics, restriction endonuclease assay was used to verify whether pcDNA3-HBV contained a complete HBV gene with correct direction. In order to verify the expression of the recombinant in mammal cells, pcDNA3-HBV was transfected into HepG2 cells with lipofectin. Positive clones were selected by G418 resistance. A high level of HBsAg was detected in culture supernatant and HBV PreS1 mRNA expression was proved by RT-PCR. These results could demonstrate that HBV can stably replicate, transcript and express in transfected host cells.
The molecular mechanisms of hepatic cell death and viral clearance during HBV infection have not been illuminated clearly yet. Studies in vitro and in vivo indicated that hepatic injury caused by HBV infection was immunity mediated[23-25]. It is considered at present that CTL was the dominate effector molecule during viral clearance, and CTL inducement was the precondition[26-28]. To simulate human hepatitis after natural HBV infection, we chose HBV complete genome transgenic BALB/c mice as the experimental model. Syngeneic BALB/c mice were undertaken gene immunity by a self-constructed HBV complete genome expression vector to induce HBV specific CTLs. Mice models of viral hepatitis B were established after transgenic mice administrated CTLs with different schemes.
There were two reasons to adopt gene immunity method. First, HBV complete genome recombinant of ayw subtype was constructed because of the transgenic mice containing HBV of the same subtype, while the main component of the present gene vaccines was HBsAg with no definite subtype. Second, generally speaking, humoral immunity could be induced by protein antigens, while cell mediated immunity could be induced by gene immunization[29-31]. There were several ways of gene transfer for gene immunity, which would influence immune effect and even determine the type of immunity. Skeletal muscle has been found to be the exclusive tissue that can efficiently ingest and express foreign genes, especially for plasmid DNA[32,33]. Because of its simpleness and convenience, it has become the dominant method for immunity. After immunized by pcDNA3-HBV once 2-3 wk, HBsAb was detected in the serum of BALB/c mice at the 4th wk. Pretreatment with 250g/L sucrose improved immune effect and induced HBV specific CTLs with a high activity.
The liver would have the most severe pathological changes in viral hepatitis B. The essential changes included degeneration and necrosis of hepatic cells, inflammatory cell infiltration, regeneration of hepatic cells, hyperplasia of fibrous tissue, and increase of serum transaminase[34,35]. HBV specific CTLs produced by immunized mice were transferred into HBV-transgenic mice. Forty-eight hours later, normal serum protein content and a high level of serum AST and ALT were detected both in negative and blank control groups. The liver stained by HE in the model group showed typical pathological changes of acute viral hepatitis B, while no pathological damage was found in the negative control group. The kidney had no pathological changes in all groups. Acute hepatitis model of mice with HBV complete genome has been established successfully by the above research project, which could mimic human hepatitis B much better because of viral replication and HBV antigens expression in the model. This study would provide a new way for anti-HBV drug screening and mechanism investigation of liver injury due to HBV infection.
Edited by Zhang JZ and Wang XL Proofread by Xu FM
| 1. | Ryu WS. Molecular aspects of hepatitis B viral infection and the viral carcinogenesis. J Biochem Mol Biol. 2003;36:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 2. | Ke WM, Ye YN, Huang S. Discriminant function for prognostic indexes and probability of death in chronic severe hepatitis B. J Gastroenterol. 2003;38:861-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Rabe C, Pilz T, Klostermann C, Berna M, Schild HH, Sauerbruch T, Caselmann WH. Clinical characteristics and outcome of a cohort of 101 patients with hepatocellular carcinoma. World J Gastroenterol. 2001;7:208-215. [PubMed] |
| 4. | Wu Jm, Lin JS, Chen BT, Zheng XM, Zhao HB, Liang KH. [Establishment and identification of highly expressing and replicating hepatitis B virus genome transgenic mouse models]. Zhonghua Ganzangbing Zazhi. 2003;11:338-340. [PubMed] |
| 5. | Wieland SF, Vega RG, Müller R, Evans CF, Hilbush B, Guidotti LG, Sutcliffe JG, Schultz PG, Chisari FV. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J Virol. 2003;77:1227-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 94] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Interleukin-18 inhibits hepatitis B virus replication in the livers of transgenic mice. J Virol. 2002;76:10702-10707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 146] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 7. | Sureau C, Romet-Lemonne JL, Mullins JI, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 289] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | Marinos G, Torre F, Chokshi S, Hussain M, Clarke BE, Rowlands DJ, Eddleston AL, Naoumov NV, Williams R. Induction of T-helper cell response to hepatitis B core antigen in chronic hepatitis B: a major factor in activation of the host immune response to the hepatitis B virus. Hepatology. 1995;22:1040-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Guidotti LG. The role of cytotoxic T cells and cytokines in the control of hepatitis B virus infection. Vaccine. 2002;20 Suppl 4:A80-A82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Liu DX. A new hypothesis of pathogenetic mechanism of viral hepatitis B and C. Med Hypotheses. 2001;56:405-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Sun J, Hou JL, Xiao L, Wang ZH, Luo KX. Analysis of three lamivudine-resistant HBV mutants with the method of restric-tion enzyme digestion and its application. Zhonghua Shiyan He Linchuang Bingdu Xue Zazhi. 2003;17:18-20. |
| 12. | Muchmore EA. Chimpanzee models for human disease and immunobiology. Immunol Rev. 2001;183:86-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Cooper A, Paran N, Shaul Y. The earliest steps in hepatitis B virus infection. Biochim Biophys Acta. 2003;1614:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Chang SF, Netter HJ, Hildt E, Schuster R, Schaefer S, Hsu YC, Rang A, Will H. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J Virol. 2001;75:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol. 2001;75:2900-2911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 96] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Babinet C, Farza H, Morello D, Hadchouel M, Pourcel C. Specific expression of hepatitis B surface antigen (HBsAg) in transgenic mice. Science. 1985;230:1160-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 112] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Chisari FV, Pinkert CA, Milich DR, Filippi P, McLachlan A, Palmiter RD, Brinster RL. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 202] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 18. | Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158-6169. [PubMed] |
| 19. | Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 428] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 20. | Ando K, Moriyama T, Guidotti LG, Wirth S, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J Exp Med. 1993;178:1541-1554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 327] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Roman M, Martin-Orozco E, Goodman JS, Nguyen MD, Sato Y, Ronaghy A, Kornbluth RS, Richman DD, Carson DA, Raz E. Immunostimulatory DNA sequences function as T helper-1-promoting adjuvants. Nat Med. 1997;3:849-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 717] [Cited by in RCA: 683] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 22. | Klinman DM, Yamshchikov G, Ishigatsubo Y. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J Immunol. 1997;158:3635-3639. [PubMed] |
| 23. | Moriyama T, Guilhot S, Klopchin K, Moss B, Pinkert CA, Palmiter RD, Brinster RL, Kanagawa O, Chisari FV. Immunobiology and pathogenesis of hepatocellular injury in hepatitis B virus transgenic mice. Science. 1990;248:361-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 266] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 24. | Tang TJ, Kwekkeboom J, Laman JD, Niesters HG, Zondervan PE, de Man RA, Schalm SW, Janssen HL. The role of intrahepatic immune effector cells in inflammatory liver injury and viral control during chronic hepatitis B infection. J Viral Hepat. 2003;10:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Kasahara S, Ando K, Saito K, Sekikawa K, Ito H, Ishikawa T, Ohnishi H, Seishima M, Kakumu S, Moriwaki H. Lack of tumor necrosis factor alpha induces impaired proliferation of hepatitis B virus-specific cytotoxic T lymphocytes. J Virol. 2003;77:2469-2476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Sobao Y, Sugi K, Tomiyama H, Saito S, Fujiyama S, Morimoto M, Hasuike S, Tsubouchi H, Tanaka K, Takiguch M. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J Hepatol. 2001;34:922-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Roh S, Lee YK, Ahn BY, Kim K, Moon A. Induction of CTL responses and identification of a novel epitope of hepatitis B virus surface antigens in C57BL/6 mice immunized with recombinant vaccinia viruses. Virus Res. 2001;73:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 28. | Street M, Herd K, Londono P, Doan T, Dougan G, Kast WM, Tindle RW. Differences in the effectiveness of delivery of B- and CTL-epitopes incorporated into the hepatitis B core antigen (HBcAg) c/e1-region. Arch Virol. 1999;144:1323-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Prugnaud JL. [DNA vaccines]. Ann Pharm Fr. 2003;61:219-233. [PubMed] |
| 30. | Wang J, Murakami T, Yoshida S, Matsuoka H, Ishii A, Tanaka T, Tobita K, Ohtsuki M, Nakagawa H, Kusama M. Predominant cell-mediated immunity in the oral mucosa: gene gun-based vaccination against infectious diseases. J Dermatol Sci. 2003;31:203-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 31. | Isaguliants MG, Petrakova NV, Mokhonov VV, Pokrovskaya K, Suzdaltzeva YG, Krivonos AV, Zaberezhny AD, Garaev MM, Smirnov VD, Nordenfelt E. DNA immunization efficiently targets conserved functional domains of protease and ATPase/helicase of nonstructural 3 protein (NS3) of human hepatitis C virus. Immunol Lett. 2003;88:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465-1468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2667] [Cited by in RCA: 2755] [Article Influence: 78.7] [Reference Citation Analysis (0)] |
| 33. | Corr M, von Damm A, Lee DJ, Tighe H. In vivo priming by DNA injection occurs predominantly by antigen transfer. J Immunol. 1999;163:4721-4727. [PubMed] |
| 34. | Myers RP, Tainturier MH, Ratziu V, Piton A, Thibault V, Imbert-Bismut F, Messous D, Charlotte F, Di Martino V, Benhamou Y. Prediction of liver histological lesions with biochemical markers in patients with chronic hepatitis B. J Hepatol. 2003;39:222-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 254] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 35. | Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641-644. [PubMed] |