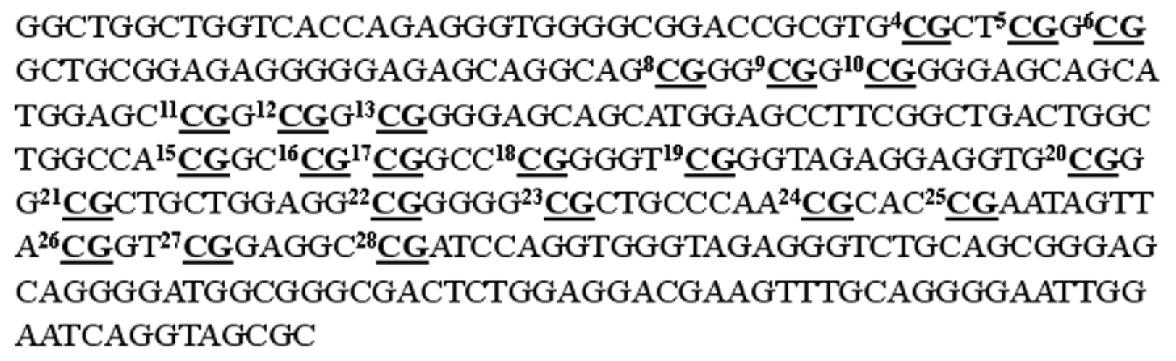Published online Dec 15, 2004. doi: 10.3748/wjg.v10.i24.3553
Revised: March 11, 2004
Accepted: March 18, 2004
Published online: December 15, 2004
AIM: Aberrant DNA methylation of CpG site is among the earliest and most frequent alterations in cancer. Several studies suggest that aberrant methylation of the CpG sites of the tumor suppressor gene is closely associated with carcinogenesis. However, large-scale analysis of candidate genes has so far been hampered by the lack of high-throughput approach for analyzing DNA methylation. The aim of this study was to describe a microarray-based method for detecting changes of DNA methylation in cancer.
METHODS: This method used bisulfite-modified DNA as a template for PCR amplification, resulting in conversion of unmethylated cytosine, but not methylated cytosine, into thymine within CpG islands of interest. Therefore, the amplified product might contain a pool of DNA fragments with altered nucleotide sequences due to differential methylation status. Nine sets of oligonucleotide probes were designed to fabricate a DNA microarray to detect the methylation changes of p16 gene CpG islands in gastric carcinomas. The results were further validated by methylation-specific PCR (MSP).
RESULTS: The experimental results showed that the microarray assay could successfully detect methylation changes of p16 gene in 18 gastric tumor samples. Moreover, it could also potentially increase the frequency of detecting p16 methylation from tumor samples than MSP.
CONCLUSION: Microarray assay could be applied as a useful tool for mapping methylation changes in multiple CpG loci and for generating epigenetic profiles in cancer.
- Citation: Hou P, Shen JY, Ji MJ, He NY, Lu ZH. Microarray-based method for detecting methylation changes of p16Ink4a gene 5’-CpG islands in gastric carcinomas. World J Gastroenterol 2004; 10(24): 3553-3558
- URL: https://www.wjgnet.com/1007-9327/full/v10/i24/3553.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i24.3553
The epigenetic event has been observed in GC-rich regions, called CpG islands, frequently located in the promoter and the first exon regions of genes. CpG island hypermethylation is closely associated with transcriptional inactivation of tumor suppressor genes, which is a common feature in human carcinomas[1]. Hypermethylated CpG islands therefore play a causal role in promoting tumor development and are useful molecular markers for cancer diagnosis and prognosis. p16, an inhibitor of the cyclin D-dependent protein kinases, is a classic tumor suppressor gene, and its inactivation is closely associated with carcinogenesis. Hypermethylation of the CpG islands of the p16 gene has been proposed as an alternative mechanism for the loss of p16 expression. p16 hypermethylation could be detected in each stage, which is consistent with the finding that aberrant methylation of p16 is a very early event in carcinogenesis[2]. Detection of promoter hypermethylation of cancer-related genes may be useful for cancer diagnosis or the detection of recurrence[3,4].
At present, several molecular biology methods are routinely used to determine the methylation status of a CpG island, such as Southern blot[5], bisulfite genomic DNA sequencing[6], restriction enzyme-PCR[7], methylation-specific PCR (MSP)[8], methylation-sensitive single nucleotide primer extension (MS-SNuPE)[9], electrochemistry[10], etc. Among these, bisulfite nucleotide sequencing is a standard technique for detailed mapping of methylated cytosine residues within a gene promoter. This meticulous method, developed by Frommer et al[6] relies on the ability of sodium bisulfite to deaminate cytosine residues into uracil in genomic DNA, whereas the methylated cytosine residues are resistant to this modification. The target DNA is then amplified by PCR with specific primers to yield fragments in which all uracil residues are converted to thymine, whereas methylated cytosine residues are amplified as cytosine. The PCR products are sequenced and the methylation status of individual CpG sites is then analyzed by comparing it with the unmodified sequences of a given promoter. Using this conventional method, many investigators have addressed the importance of promoter CpG hypermethylation in the regulation of specific gene transcription in cancer[11-14]. The method, which requires cloning and sequencing of individual inserts, can be labor intensive and is restricted to the evaluation of DNA methylation on a gene-by-gene basis. Such an approach has given researchers a limited picture of complex epigenetic alterations in cancer. Clearly, it is of great importance to establish novel, reliable and high-throughput methods for the methylation detection of earlier cancer diagnosis.
For this purpose, considerable advances have been made in hybridization-based microarray technology for genome-wide analysis of gene mutations and single nucleotide polymorphisms[15-17]. In this new approach, oligonucleotides are arrayed on solid supports known as probes, and the labeled complex DNA mixtures to be interrogated are known as targets[18]. Recently, we developed an oligonucleotide microarray to analyze methylation patterns of several adjacent CpG sites[19]. The DNA microarray can successfully map the methylation pattern of p16Ink4a gene. However, it can not be used to quantify the methylation level of promoter region of gene. Gitan et al[20] have developed a methylation specific oligonucleotide (MSO) microarray to analyze methylation of human estrogen receptor (ER) α gene. The targets were derived from PCR products of bisulfite-modified DNA, whereas the probes used a series of arrayed oligonucleotides that can discriminate between converted and unconverted nucleotides, that is, unmethylated and methylated cytosines, at CpG sites. The MSO microarray is a novel and powerful tool for determining the methylation level in multiple CpG island loci and for generating epigenetic profiles in cancer.
The aim of this study was to use oligonucleotides microarray method to analyze methylation changes of p16 gene CpG islands in gastric carcinomas. A 336 bp segment was selected in the 5’ untranslated region and the first exon of the p16 gene, as the investigated target, which contain 32 CpG sites. Nine sets of oligonucleotide probes were designed to test 23 CpG sites within the island. Here, we described the oligonucleotide microarray procedure and its application for analyzing the methylation changes of p16 gene CpG islands in gastric tumor and corresponding normal tissues.
Eighteen gastric tumor and corresponding normal tissues were obtained from Gulou Hospital (Nanjing, China). Genomic DNA was isolated by standard methods using proteinase K digestion and phenol/chloroform extraction.
Bisulfite processing of DNA was performed as described by Frommer et al[6] and the modifications introduced by Clark et al[21]. Briefly, 1 μg of genomic DNA was digested by EcoR I and denatured in 0.35 mol/L NaOH at 37 °C for 20 min. Bisulfite reaction was carried out in 3.2 mol/L sodium bisulfite and 0.5 mmol/L hydroquinone (Sigma Chemical Co., USA) at 55 °C for 16-24 h. DNA was recovered by a desalting column (DNA Clean-Up System, Promega Inc., USA) and desulphonated in 0.2 mol/L NaOH at 37 °C for 15 min, neutralized by ammonium acetate, alcohol precipitated, dried and then dissolved in 30 μL of deionized water. After bisulfite processing, all unmethylated cytosine residues converted to uracil, whereas the methylated ones remained unchanged. For bisulfite genomic sequencing the sense strand of a 336 bp fragment of the p16 gene 5’-CpG island corresponding to nucleotides -128 to +208 relatively to the transcription start site[22] was amplified with primers which did not contain cytosine in a CpG context and consequently annealed to the methylated status of the island. The sequence of forward primer is 5’-AAAGAGGAGGGGTTGGTTGGTTATTA-3’ and that of backward primer is 5’-TACCTAATTCCAATTCCCCTACAAACT-3’. (Forward primer position 5-30 and reverse primer position 310-336 in GenBank accession number U12818). PCR-reaction was performed in a buffer containing 10 mmol/L Tris-HCl (pH9.0), 50 mmol/L KCl, 1 g/L Triton X-100, 50 g/L DMSO, 1.75 mmol/L MgCl2, 0.2 mmol/L of each dNTP and 1 μL bisulphite treated DNA. Amplification conditions were as follows: at 95 °C for 5 min; followed by 35 cycles, each at 95 °C for 1 min, at 62 °C for 1 min, and extension at 72 °C for 30 s, and ended with an extension at 72 °C for 7 min and quickly chilled to 4 °C on a PTC-225 thermocycler (MJ Research). PCR products were gel purified and cloned into the pMD18-T vector according to the manufacturer’s instructions (TAKARA). Part of the same PCR products was fluorescently labeled later for MSO microarray analysis. Plasmid DNA from 30 positive recombinant clones was isolated, and inserts were sequenced on an automated sequence analyzer (ABI377A, Applied Biosystem Inc., USA).
Nine sets of paired oligonucleotides used in this study were designed to include two or three CpG sites of the p16 CpG island to be interrogated (Table 1). These oligonucleotides were specific to the bisulfite-modified sequence of portion of the p16 CpG island. Each was synthesized with amino-linked C6 [NH2(CH2)6] linker attached to its 5’ end. These oligonucleotides were suspended in sodium carbonate buffer (0.1 mol/L, pH9.0) to a final concentration of 80 μmol/L. Approximately, 1 nL (0.05-0.1 pmole) of each oligonucleotide was printed on the aldehyde-coated glass slides (DAKO) using a PixSys5500 microarrayer (Cartesian Technology Inc). After printed, the glass slides were incubated in a humid chamber at room temperature overnight, and then at 37 °C for 2 h. The slides were washed thoroughly in 1 g/L SDS to remove unbound oligonucleotides. After further treatment with a NaBH4 solution for 15 min, the slides were ready for hybridization. For target labeling, PCR products of bisulfite-treated DNA were labeled with Cy3-dCTP (Amersham Pharmacia) by terminal transferase (TAKARA). The unincorporated dCTP was removed by passing the labeled target through a micro-Biospin column (Bio-Rad). The labeled products were resuspended in hybridization solution (1:3 dilution v/v). Then the mixture was denatured at 95 °C for 5 min, cooled to room temperature, and applied to the DNA microarray slides. Microarray hybridization was conducted in a moist hybridization chamber under a cover slip at 42 °C for 2 h. After hybridization, the slide was rinsed and washed at room temperature with 2 × SSC-1 g/L SDS and 0.1 × SSC-1 g/L SDS for a total of 15 min, respectively, and then dried by centrifugation at 600 rm for 5 min.
| p16 CpG sites | Oligonucleotide sequences | Tm (°C) | |
| #4-6 | M: | 5’-NH2-(T)10-CAACCGCCGAACGCAC-3’ | 56 |
| U: | 5’-NH2-(T)10-CAACCACCAAACACAC-3’ | 50 | |
| #8-10 | M: | 5’-NH2-(T)10-CAACCGCCGAACGCAC-3’ | 61 |
| U: | 5’-NH2-(T)10-CCACCACCCACTACCTA-3’ | 53 | |
| #11-13 | M: | 5’-NH2-(T)10-CCGCCGCCGACTCCAT-3’ | 61 |
| U: | 5’-NH2-(T)10-CCACCACCAACTCCAT-3’ | 53 | |
| #15-17 | M: | 5’-NH2-(T)10-AACCGCGACCGTAACCAA-3’ | 58 |
| U: | 5’-NH2-(T)10-AACCACAACCATAACCAA-3’ | 50 | |
| #18,19 | M: | 5’-NH2-(T)10-TCTACCCGACCCCGAACC-3’ | 60 |
| U: | 5’-NH2-(T)10-TCTACCCAACCCCAAACC-3’ | 55 | |
| #20,21 | M: | 5’-NH2-(T)10-AACAACGCCCGCACCTC-3’ | 57 |
| U: | 5’-NH2-(T)10-AACAACACCCACACCTC-3’ | 50 | |
| #22,23 | M: | 5’-NH2-(T)10-ACAACGCCCCCGCCTC-3’ | 59 |
| U: | 5’-NH2-(T)10-ACAACACCCCCACCTC-3’ | 52 | |
| #24,25 | M: | 5’-NH2-(T)10-AACTATTCGATACGTTAAAC-3’ | 48 |
| U: | 5’-NH2-(T)10-AACTATTCAATACATTAAAC-3’ | 39 | |
| #26-28 | M: | 5’-NH2-(T)10-ATCGACCTCCGACCGTAAC-3’ | 55 |
| U: | 5’-NH2-(T)10-ATCAACCTCCAACCATAAC-3’ | 49 |
DNA microarray slide was scanned with ScanArray Lite microarray analysis systems (A Packard BioScience Company, USA) after the above treatment. Images acquired by the scanner were analyzed with software Genepix Pro 3.0. Each spot was defined by the positioning of a grid of circles over the array image. For each fluorescent image, the average pixel intensity within each circle was determined and a local background using mean pixel intensity was computed for each spot. The net signal was determined by subtraction of this local background from the mean average intensity for each spot. The data generated by the software were exported in a spreadsheet format and processed using Microsoft Excel. Statistical analyses were conducted using Origin 5.0 software.
The 5’-CpG island regions of the p16 gene were amplified with primers for methylated and unmethylated DNA, respectively. Primer pairs described in Table 2[8] were synthesized and purified by Shengyou Inc (Shanghai, China). PCR amplification was performed in a buffer containing 10 mmol/L Tris-HCl (pH9.0), 50 mmol/L KCl, 1 g/L Triton X-100, 50 g/L DMSO, 1.75 mmol/L MgCl2, 0.2 mmol/L of each dNTP and 1 μL bisulfite treated DNA in a final volume of 30 μL. The amplification was carried out for 35 cycles (30 s at 95 °C, 30 s at the annealing temperature listed in Table 2, and 30 s at 72 °C), followed by a final 4-min extension at 72 °C and quickly chilled to 4 °C on a PTC-225 thermocycler (MJ Research). Products amplified with both types of primers were examined on 10 g/L agarose gel.
| Primer sets | Sequences (5’→3’) | Size bp | Annealing temp (°C) |
| MS | TTATTAGAGGGTGGGGCGGATCGC | 234 | 65 |
| MA1 | CCACCTAAATCGACCTCCGACCG | ||
| US | TTATTAGAGGGTGGGGTGGATTGT | 234 | 60 |
| UA1 | CCACCTAAATCAACCTCCAACCA | ||
| MS | TTATTAGAGGGTGGGGCGGATCGC | 150 | 65 |
| MA2 | GACCCCGAACCGCGACCGTAA | ||
| US | TTATTAGAGGGTGGGGTGGATTGT | 151 | 60 |
| UA2 | CAACCCCAAACCACAACCATAA |
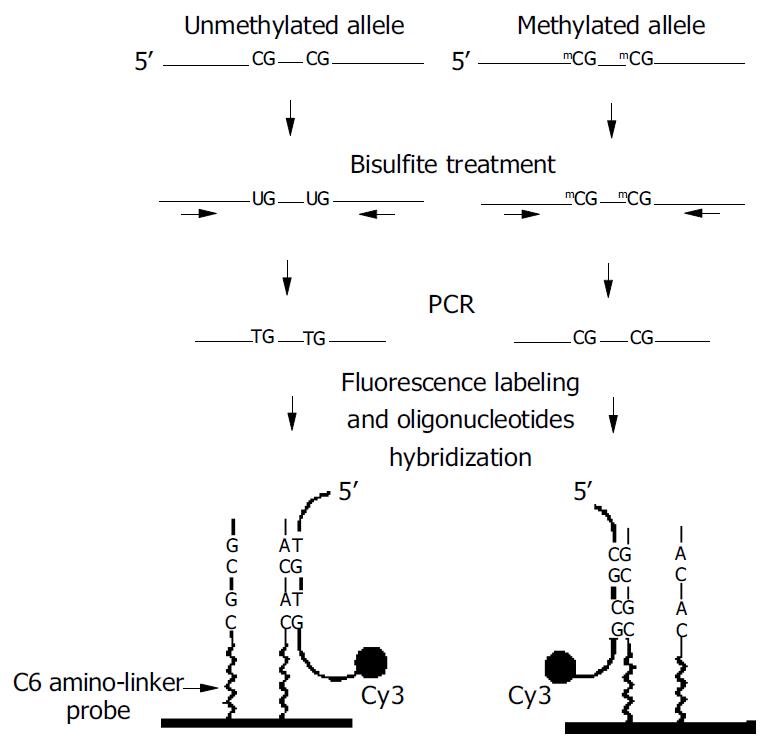
Figure 1 outlines the MSO strategy for DNA methylation analysis. Test DNA samples were bisulfite-modified, PCR amplified products contained pools of DNA fragments with altered nucleotide sequences due to their differential methylation status. As shown, the unmethylated allele of a given DNA sequence was expected to have the unmethylated cytosine of the tested CpG sites converted to thymine, whereas these CpG sequences remained unchanged in the methylated allele. Target DNA was then hybridized to arrayed oligonucleotides probes specifically designed to discriminate between converted and unconverted nucleotides at these CpG sites.
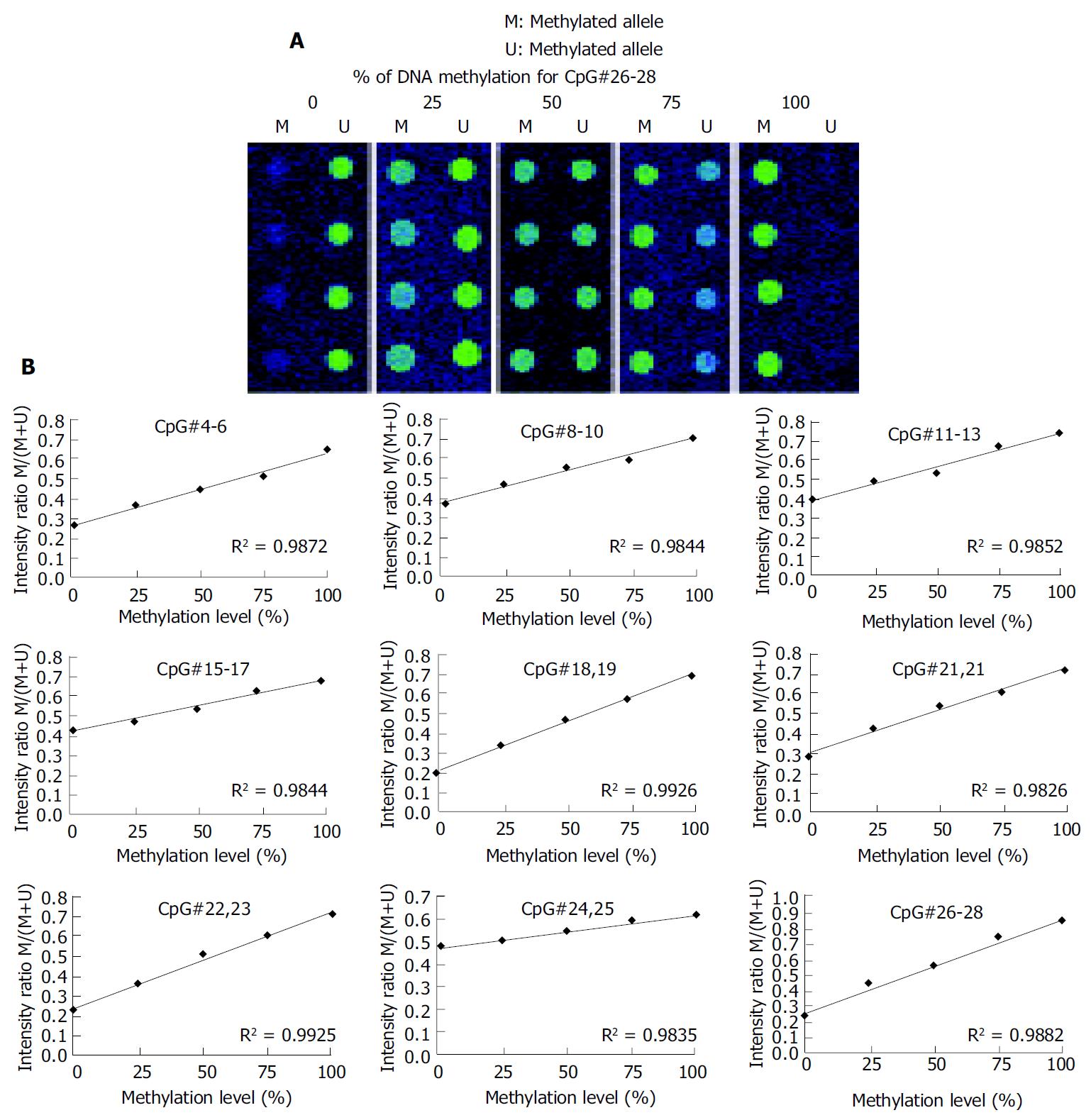
A 336 bp segment was selected in the 5’ untranslated region and the first exon of the p16 gene, as the investigated target, which contains 32 CpG sites (Figure 2). Nine sets of oligonucleotide probes were designed to test 23 CpG sites within the island, each set contained a pair of methylated and unmethylated oligonucleotides for interrogating 2 or 3 CpG sites in close proximity (Table 1). First, control DNA targets were used to test the accuracy and reproducibility of probes designed for microarray hybridization. We selected fully methylated and unmethylated ones as positive and negative controls from 36 positive recombinant clones. The positive control generated in this way remained 100% cytosine in the tested CpG sites, whereas the negative control had all cytosine residues converted into thymine in the tested CpG sites. Next, a series of microarray hybridization were performed with mixtures of Cy3 labeled positive and negative DNA targets at different proportions representing 0%, 25%, 50%, 75%, and 100% of DNA methylation to test the linearity of the protocol. An example of the microarray analysis for CpG#26-28 is shown in Figure 3A. The average intensity of hybridization signals from the four replicate spots for the methylated (M) and unmethylated (U) alleles was then derived and used to calculate the intensity ratio of M/(M + U). In this case, a linear relationship (R2 = 0.9882) was established, showing that the increase in DNA methylation was proportional to the increase in intensity ratios in the control samples. The result suggested that this set of oligonucleotide probes was optimal for the detection of methylation changes at CpG#26-28. This approach was used to test other oligonucleotide probes and to generate a set of standards for the calibration of DNA methylation changes in the test samples (Figure 3B). We noticed that the regression line for CpG#24, 25 was much higher on the Y-axis than the rest of the CpG sites. Moreover, its slope was much lower than the others. This higher nonspecific hybridization was likely due to the lower melting temperature of the unmethylated probe. An oligonucleotide sequence such as this would result in the compression of the usable scale and make the assessment of methylation status a little more challenging.
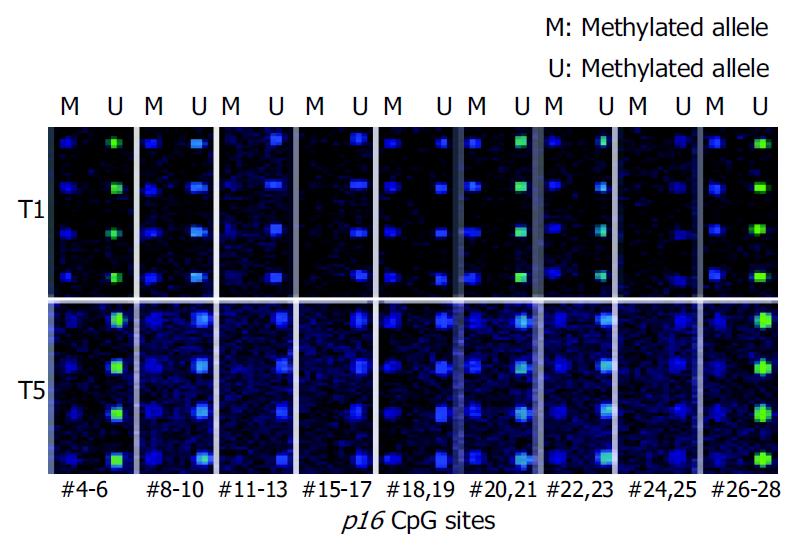
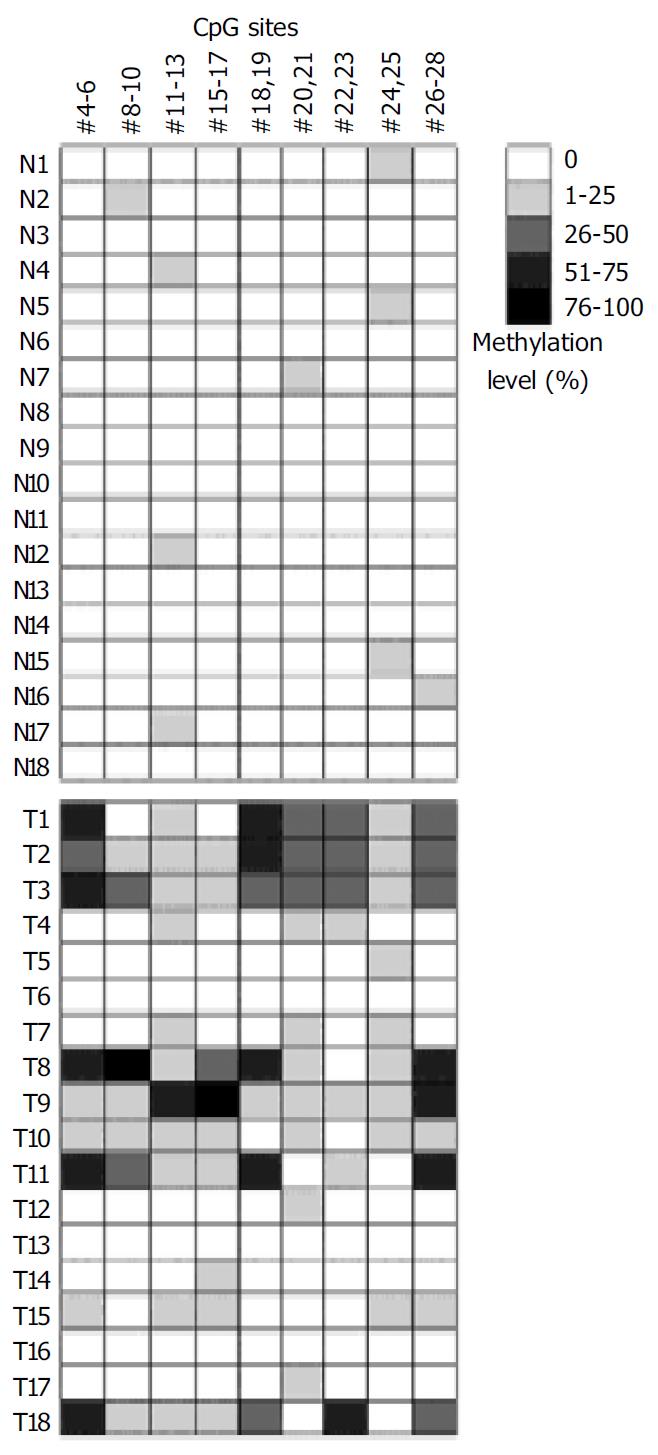
Microarray assay was used to analyze the methylation status of 18 gastric tumor and corresponding normal tissues. Figure 4 shows the representative examples of microarray results. By use of the standard curves derived from the aforementioned calibration controls, no methylation was detected in the normal tissues. Extensive methylation of the p16 CpG island was observed in 7 of 18 gastric tumor tissues (T1, T2, T3, T8, T9, T11, and T18), a modest degree of methylation was found in T10 and T15, whereas little or no methylation was seen in others (Figure 5).
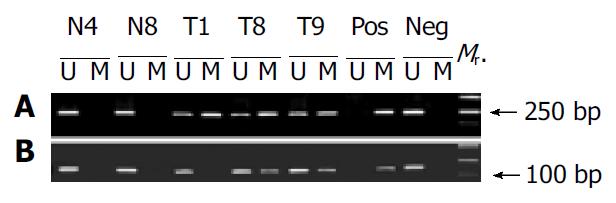
To further validate the microarray findings in gastric tumors, methylaton-specific PCR (MSP) was conducted in 18 tumor and corresponding normal tissues. The primers MA1 and MA2 for MSP included CpG#16-19 and CpG#26-28 sites, respectively. Therefore, CpG#15-17 and CpG#26-28 sites were used for this conformation. A representation of the MSP analysis is shown in Figure 6. By use of this approach, two normal tissues were completely unmethylated (N4 and N8). MSP results of the 18 gastric tumor tissues completely matched with microarray results. Interestingly, the MSP results indicated that the methylation was not detected in T1 when the amplification was performed with primers MS and MA2, whereas the methylation could be found in T1 when the amplification was performed with primers MS and MA1 (Figure 6). A most possible reason was that some CpG sites were not methylated in the region of primer MA2. Figure 5 indicates that CpG#15-17 sites had no methylation in T1, whereas CpG#26-28 sites had 47% methylation in T1, which were consistent with the MSP results.
The above results indicated that microarray assay could potentially increase the frequency of detecting p16 methylation from tumor samples than MSP. MSP was a simple, sensitive, and specific method for determining the methylation status of virtually any CpG-rich region. However, methylation could not be detected when some CpG sites were not methylated in region of MSP primers. The issue could be easily overcome using microarray assay. Furthermore, microarray assay could estimate which CpG sites (or CpG-rich region) were easily methylated in certain tumors.
In this study, we have applied a microarray method to a comprehensive analysis of DNA methylation. The results indicated that the microarray was successfully used to map methylated CpG sites within the p16 gene CpG islands in clinical samples. The derived methylation information for gastric tumor and corresponding normal tissues was assessed quantitatively and independently validated by methylation-specific PCR.
This microarray-based analysis of DNA methylation is expected to provide new tools for research in this field. At present, most methylation assays have been limited to analyzing CpG islands of a few known genes and are restricted in throughput for a genome-wide analysis. Until recently, Gitan et al[20] have developed a novel technique called MSO microarray that combines bisulfite DNA assay and oligonucleotides microarray for analysis of DNA methylation. The MSO microarray potentially allows rapid screening of multiple CpG sites in many gene promoters. CpG island hypermethylation has been reported to be linked to the silencing of more than 100 cancer-related genes. A DNA microarray can be generated to contain hundreds of oligonucleotides designed to discriminate between methylated and unmethylated sequences in these gene promoters. Bisulfite-treated genomic DNA from each of these loci can be amplified from investigated samples in a 96-well format to generate multiple targets for oligonucleotides hybridization.
As with other oligonucleotide microarrays, cross-hybridization between imperfect-match probes and targets could be observed. In addition, some probes might inherently diminish hybridization signals, probably due to decreased duplex stability of targets and probes[18]. Through careful data analysis, Gitan and his colleagues considered that cross-reactivity might also increase when oligonucleotide probes were designed to query methylation differences in one single CpG site. The issue is easily overcome by designing probes to include two or more CpG sites. This design consideration may limit the MSO assay’s ability to detect methylation changes in single CpG sites. Adorjan et al[23] have developed a microarray-based assay that can analyze methylation changes of single CpG sites. Several hundred CpG sites were screened in 76 samples from four different human tumor types and corresponding healthy controls. The results demonstrated that the microarray could be applied as a powerful tool to the assessment of selected CpG dinucleotides and quantification of methylation at each site. As shown in this study, the use of a simple control system could test the accuracy and reproducibility of the probes designed for microarray hybridization. This control system can also be used to calibrate the levels of methylation changes detected in the investigated samples by microarray assay.
In summary, microarray assay can be readily used to high-throughput analysis of DNA methylation. It will contribute significant information to our understanding of CpG island methylation in cancer.
Edited by Wang XL Proofread by Zhu LH and Xu FM
| 1. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1307] [Cited by in RCA: 1210] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 2. | Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891-11896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 700] [Cited by in RCA: 661] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 3. | Valenzuela MT, Galisteo R, Zuluaga A, Villalobos M, Núñez MI, Oliver FJ, Ruiz de Almodóvar JM. Assessing the use of p16(INK4a) promoter gene methylation in serum for detection of bladder cancer. Eur Urol. 2002;42:622-628; discussion 622-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67-70. [PubMed] |
| 5. | Bickle TA, Krüger DH. Biology of DNA restriction. Microbiol Rev. 1993;57:434-450. [PubMed] |
| 6. | Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827-1831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2250] [Cited by in RCA: 2264] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 7. | Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808-811. [PubMed] |
| 8. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4247] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 9. | Kuppuswamy MN, Hoffmann JW, Kasper CK, Spitzer SG, Groce SL, Bajaj SP. Single nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis genes. Proc Natl Acad Sci USA. 1991;88:1143-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 114] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Hou P, Ji M, Ge C, Shen J, Li S, He N, Lu Z. Detection of methylation of human p16(Ink4a) gene 5'-CpG islands by electrochemical method coupled with linker-PCR. Nucleic Acids Res. 2003;31:e92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Hiltunen MO, Alhonen L, Koistinaho J, Myöhänen S, Pääkkönen M, Marin S, Kosma VM, Jänne J. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int J Cancer. 1997;70:644-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Stirzaker C, Millar DS, Paul CL, Warnecke PM, Harrison J, Vincent PC, Frommer M, Clark SJ. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229-2237. [PubMed] |
| 13. | Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 170] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730-3740. [PubMed] |
| 15. | Ahrendt SA, Halachmi S, Chow JT, Wu L, Halachmi N, Yang SC, Wehage S, Jen J, Sidransky D. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci USA. 1999;96:7382-7387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 164] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Favis R, Barany F. Mutation detection in K-ras, BRCA1, BRCA2, and p53 using PCR/LDR and a universal DNA microarray. Ann N Y Acad Sci. 2000;906:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Wen WH, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, Press MF. Comparison of TP53 mutations identified by oligonucleotide microarray and conventional DNA sequence analysis. Cancer Res. 2000;60:2716-2722. [PubMed] |
| 18. | Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 314] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 19. | Hou P, Ji M, Liu Z, Shen J, Cheng L, He N, Lu Z. A microarray to analyze methylation patterns of p16Ink4a gene 5'-CpG islands. Clin Biochem. 2003;36:197-202. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 189] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1369] [Cited by in RCA: 1387] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 22. | Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859-867. [PubMed] |
| 23. | Adorján P, Distler J, Lipscher E, Model F, Müller J, Pelet C, Braun A, Florl AR, Gütig D, Grabs G. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 185] [Article Influence: 8.0] [Reference Citation Analysis (0)] |