Published online Dec 1, 2004. doi: 10.3748/wjg.v10.i23.3514
Revised: February 22, 2004
Accepted: April 13, 2004
Published online: December 1, 2004
AIM: To set up a real-time fluorescent quantitative reverse transcription-polymerase chain reaction (RT-PCR) assay, to detect human telomerase reverse transcriptase (hTERT) messenger RNA in gastric carcinomas, and to evaluate quantitative determination of hTERT mRNA in the diagnostic value of gastric carcinomas, and to analyze the correlation between the expression level of hTERT mRNA and clinicopath-ological parameters in patients with gastric cancer.
METHODS: A real-time quantitative RT-PCR (RQ-PCR) based on TaqMan fluorescence methodology and the LightCycler system was used to quantify the full range of hTERT mRNA copy numbers in 35 samples of gastric carcinomas and corresponding adjacent non-cancerous tissues. The normalized hTERT (NhTERT) was standardized by quantifying the number of GAPDH transcripts as internal control and expressed as 100 × (hTERT/GAPDH) ratio. Variables were analyzed by the Student’s t-test, χ2 test and Fisher’s exact test.
RESULTS: NhTERT from gastric carcinomas and corresponding adjacent non-cancerous tissues was 6.27 ± 0.89 and 0.93 ± 0.18, respectively (t = 12.76, P < 0.001). There was no significant association between gastric cancer hTERT mRNA expression level and patient’s age, gender, tumor size, location and stage (PTNM), but a significant correlation was found between hTERT mRNA expression level in gastric carcinomas and the degree of differentiation.
CONCLUSION: Quantitative determination of hTERT mRNA by RQ-PCR is a rapid and sensitive method. hTERT might be a potential biomarker for the early detection of gastric cancer.
- Citation: Hu LH, Chen FH, Li YR, Wang L. Real-time determination of human telomerase reverse transcriptase mRNA in gastric cancer. World J Gastroenterol 2004; 10(23): 3514-3517
- URL: https://www.wjgnet.com/1007-9327/full/v10/i23/3514.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i23.3514
Telomerase is a reverse transcriptase that adds telomeric repeats to chromosomal ends to compensate for sequence loss during DNA replication. Telomerase activity has been detected in about 85% of human cancer samples and is associated with cell immortalization and the acquisition of malignancy, but most normal tissues have low or no telomerase activity[1]. Telomerase is one of the most widespread tumor markers at present. To date, the main assay to detect telomerase activity is telomere repeat amplification protocol (TRAP). TRAP is a qualitative or semi-quantitative assay, and can not accurately exhibit telomerase expression level, and needs functional ribonucleoproteins, including both reverse transcriptase activity and undegraded RNA. The presence of telomerase inhibitors, Taq polymerase inhibitors, proteases, or RNases in tissue extract may influence its detection and subsequently lower its sensitivity. With the cloning of both genes coding for human telomerase RNA (hTR) and human telomerase reverse transcriptase (hTERT), hTERT becomes the catalytic subunit of telomerase and is a rate-limiting determinant of the enzymatic activity of human telomerase, and only the expression of hTERT is closely associated with telomerase activity[2-5], whereas the expression of hTR is widespread. The close relationship between hTERT mRNA expression and telomerase activity suggests that quantification of the mRNA expression of the hTERT gene could be used as an alternative to measure telomerase activity. In this study, we used real-time quantitative reverse transcription-polymerase chain reaction (RQ-PCR) to detect and quantify hTERT mRNA in samples of gastric carcinoma and corresponding non-cancerous tissues, and to evaluate the quantitative determination of hTERT mRNA in the diagnostic value of gastric carcinomas, and to analyze the correlation between the expression level of hTERT mRNA and clinicopathological parameters in patients with gastric cancer.
We analyzed tissues (gastric cancer and corresponding non-cancerous tissues) from surgically removed primary gastric cancer in Union Hospital and Tongji Hospital of Tongji Medical College of Huazhong University of Science and Technology from October 2002 to May 2003. All patients (25 males and 10 females, mean age 55.2 years, range 34-73 years) were at initial presentation and had no radiotherapy or chemotherapy history before surgery. All samples were examined histopathologically to confirm the diagnosis. Control tissues were the corresponding non-cancerous mucosa from the stomach of cancer patients, and excised beyond 5 cm from neoplastic lesions. Samples were stored at -80 °C until further analysis.
TRIzol was the product of Omega. The reagents used for reverse transcription were purchased from Promega. The reagents used for PCR and PCR product purification, glyceraldehydes 3-phosphate dehydrogenase (GAPDH) quantification, and the primers and TaqMan probe of hTERT were all purchased from Shanghai Shenyou Company. Both T4 DNA ligase and PMD18-T vector were the products of TaKaRa. The other chemical reagents used in this study were ACS reagents of China. The fluorescent quantitative PCR instrumentation was the LightCycler system of Roche.
We used a RQ-PCR assay based on TaqMan fluorescence methodology to quantify the full range of hTERT mRNA copy numbers[6,7]. This method used a dual-labeled nonextendable oligonucleotide hydrolysis (TaqMan) probe in addition to the two amplification primers. The probe contained 6-carboxy-fluorescein (FAM) as a fluorescent reporter dye, and 6-carboxytetramethyl-rhodamine (TAMRA) as a quencher for its light emission spectrum. During the extension phase of PCR, the probe hybridized to the target sequence and was then cleaved due to the 5’ to 3’ exonuclease activity of Taq polymerase. The increase in the fluorescence signal of the reporter was proportional to the amount of specific PCR products, providing highly accurate and reproducible quantification. The number of PCR cycles to reach the fluorescence threshold was the cycle threshold (Ct). The Ct value for each sample was proportional to the log of the initial amount of input cDNA. By plotting the Ct value of an unknown sample on the standard curve, the amount of target sequences in the sample could be calculated.
To normalize the hTERT mRNA expression for sample-to-sample differences in RNA input, RNA quality, and reverse transcriptase efficiency, we amplified the housekeeping gene GAPDH. According to each standard curve, we got the copy numbers of GAPDH and hTERT, respectively. The ratio between copy numbers of hTERT and GAPDH represented the normalized hTERT (NhTERT) for each sample and could be compared with that of other samples[8].
NhTERT = (hTERT mRNA copiessample/GAPDH mRNA copiessample) × 100.
According to GAPDH quantification reagents and the reference[9], the nucleotide sequences of oligonucleotide TaqMan probes and primers are shown in Table 1.
| Gene and oligonucleotide | Sequence | PCR product size (bp) |
| GAPDH | ||
| Upper primer | 5’-GAAGGTGAAGGTCGGAGTC-3’ | |
| Lower primer | 5’-GAAGATGGTGATGGGATTTC-3’ | 226 |
| Probe | 5’-(FAM) CAAGCTTCCCGTTCTCAGCC (TAMRA)-3’ | |
| hTERT | ||
| Upper primer | 5’-TGACACCTCACCTCACCCAC-3’ | |
| Lower primer | 5’-CACTGTCTTCCGCAAGTTCAC-3’ | 95 |
| Probe | 5’-(FAM) ACCCTGGTCCGAGGTGTGTCCCTGA (TAMRA)-3’ |
According to the manufacturer’s instructions, total RNA from frozen tumor and corresponding non-cancerous tissue specimens was isolated by disruption of 50-100 mg tissues in 1 mL of TRIzol. RNA was quantified spectrophotometrically, and its quality was determined by agarose gel electrophoresis and ethidium bromide staining. Only samples that were not degraded and showed clear 18 S and 28 S bands under ultraviolet light were used for real-time RT-PCR.
All samples were denatured for 10 min at 60 °C to melt secondary structure with the template and cooled immediately on ice for 5 min to prevent secondary structure from reforming. Total RNA (1-2 µg) was reverse transcribed in a total volume of 25 µL containing 1 × RT buffer (Promega), 200 U of Moloney murine leukemia virus Reverse Transcriptase (M-MLV RT) (Promega), 20 U of RNasin (Promega), 0.2 µg random primer (Promega) and 1 mmol/L deoxynucleotides. The reaction was performed for 10 min at 25 °C, for 60 min at 42 °C and for 10 min at 70 °C. cDNA was stored at -20 °C until use.
Pure hTERT fragments from classical RT-PCR were joined to PMD18-T vector by T4 DNA ligase, resulting in recombinant plasmid PMD18-hTERT. The recombinant plasmids were confirmed in including hTERT target fragments by sequencing, and extracted and purified. The recombinant plasmid DNAs at 107 copies/mL were stored at -20 °C until use. GAPDH standard template was from the GAPDH quantification reagents.
All PCR reactions were performed using the LightCycler System (Roche Diagnostics, Switzerland) in a total volume of 20 µL containing 1 ×Taq polymerase buffer, 4 mmol/L MgCl2, 200 µmol/L deoxynucleotides, 300 nmol/L each primer, 150 nmol/L probe, 1 U Taq polymerase and 20 ng cDNA. Water instead of cDNA template was used for the negative controls. Both GAPDH and hTERT amplification were done in duplicate for each sample. The thermal cycling conditions were 5 min at 94 °C, followed by 40 cycles, each at 94 °C for 15 s and at 60 °C for 1 min for GAPDH or at 65 °C for 1 min for hTERT (two-step PCR). The number of GAPDH and hTERT transcripts in samples was calculated with the LightCycler software, using these standard curves.
Data were expressed as mean ± SD. The Student t-test, χ2 test and Fisher’s exact test were used in this study. P < 0.05 was considered statistically significant.
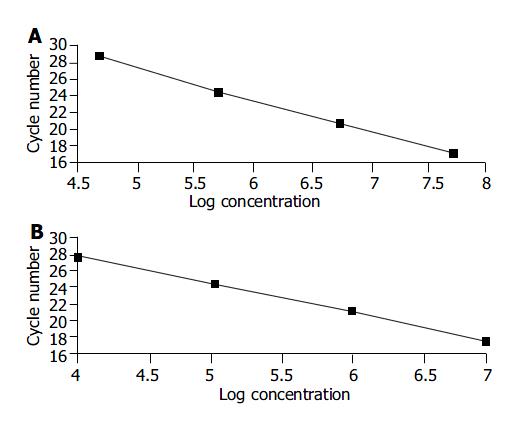
We used different concentrations of GAPDH and hTERT standard templates including 104, 105, 106 and 107 copies/mL to perform quantitative PCR and calculate the standard curves, respectively. The standard curves were: CtGAPDH = -3.86 log (GAPDH copies) + 46.60; and CthTERT = -3.42 log (hTERT copies) +37.31. The correlation coefficients were both -1.00 (Figure 1A, B).
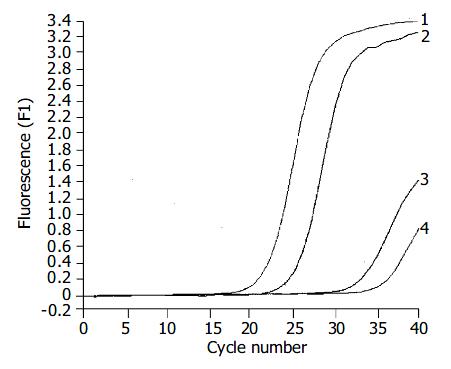
The expression of hTERT mRNA in gastric carcinomas was analyzed by a highly sensitive RQ-PCR assay. HTERT mRNA expression was detectable not only in all 35 gastric carcinomas, but also in the corresponding non-cancerous gastric tissues (Figure 2). For each experimental sample, the amount of hTERT and GAPDH was determined from the appropriate standard curve. Then, hTERT amount was divided by the GAPDH amount to obtain NhTERT. NhTERT was significantly higher (t = 12.76, P < 0.001) in tumor tissues (6.27 ± 0.89) than in the corresponding non-cancerous tissues (0.93 ± 0.18).
The data obtained by RQ-PCR were analyzed in relation to clinicopathological parameters of the patients. All 35 corresponding non-cancerous gastric tissue samples expressed low detectable NhTERT, ranging from 0.06 to 2.11. In the 35 gastric carcinomas, NhTERT varied greatly, ranging from 0.28 to 90.55. NhTERT was categorized as low or high, using a cutoff at 5.39, which was the value that represented the median value of the expression distribution. Thus, 17 tumors (48.6%) showed low hTERT expression, whereas 18 tumors (51.4%) had high hTERT expression. We observed the statistical links between hTERT mRNA expression levels and the degree of differentiation (P = 0.007). Seventy-three point six percent (14 of 19) of poorly differentiated and undifferentiated tumors showed high NhTERT expressions, whereas 75% (12 of 16) of well and moderately differentiated tumors exhibited low NhTERT expressions. Poorly differentiated and undifferentiated tumors had higher hTERT mRNA expression levels than well and moderately differentiated tumors. No relationship was found between hTERT mRNA expression level and patients’ age, gender, tumor size, tumor location and stage (pTNM). The results obtained are shown in Table 2.
| Parameter | n | NhTERT | P | |
| Low (n = 17) | High (n = 18) | |||
| Age (yr) | ||||
| < 50 | 13 | 7 | 6 | |
| ≥ 50 | 22 | 10 | 12 | 0.73 |
| Gender | ||||
| Male | 25 | 13 | 12 | |
| Female | 10 | 4 | 6 | 0.71 |
| Size of diameter (cm) | ||||
| < 5 | 19 | 11 | 8 | |
| ≥ 5 | 16 | 6 | 10 | 0.31 |
| Location | ||||
| Cardia | 14 | 8 | 6 | |
| Body and antrum Degree of differentiation | 21 | 9 | 12 | 0.50 |
| Well and moderately differentiated | 16 | 12 | 4 | |
| Poorly differentiated and undifferentiated | 19 | 5 | 14 | 0.007 |
| Lymph node metastasis | ||||
| N0 | 8 | 4 | 4 | |
| N1 | 13 | 8 | 5 | |
| N2 | 14 | 5 | 9 | 0.40 |
| Depth of invasion | ||||
| T1 and T2 | 24 | 13 | 11 | |
| T3 and T4 | 11 | 4 | 7 | 0.43 |
| Metastasis | ||||
| M0 | 20 | 12 | 8 | |
| M1 | 15 | 5 | 10 | 0.17 |
We used RQ-PCR based on TaqMan methodology for the accurate quantification of hTERT mRNA expression in gastric carcinomas. This assay has several marked advantages over the TRAP assay for cancer detection. It needs only a 95-bp fragment of hTERT mRNA, making the assay less sensitive to RNase activity and insensitive to proteases and protein inhibitors. The assay output is numerical rather than qualitative, allowing appropriate diagnostic statistics to be applied. It uses endogenous controls (GAPDH in this study) which allow correction for parameters like RNA input, RNA degradation, or RT inhibitors. Real-time PCR makes RNA quantification much more precise and reproducible, based on Ct values established in the early exponential phase of the PCR reaction (when none of the reagents is rate-limiting) rather than end point quantification of the amount of accumulated PCR product. It does not require post-PCR sample handling and the closed-tube method minimizes the risk for cross-contamination. These suggest that quantitative determination of hTERT mRNA by RQ-PCR is a powerful method to investigate the telomerase status and superior in specificity and sensitivity to the evaluation of telomerase activity by the TRAP assay. In addition, we used recombinant hTERT plasmid DNA to calculate the external standard curves, and found it was better for the quantification of mRNA than the recombinant RNA calibrator or endogenous standards[10,11]. Theoretically, the slope of the standard curve should be -3.3 if 10-fold dilutions are used, but in practice a slope between -3.0 and -3.9 is probably acceptable as long as the correlation coefficient is > 0.95. The standard curves we obtained in this study had acceptable slopes and correlation coefficients.
In this study, the level of hTERT mRNA estimated with RQ-PCR procedure was the average amount of transcripts in a whole tissue sample and mainly depended on the number of hTERT-positive cells present in the tissue. hTERT mRNA was detected in 100% of gastric carcinoma RNAs, and also in all the corresponding non-cancerous gastric mucosa RNAs. The highest NhTERT was detected in gastric cancer, whereas the lowest NhTERT was found in non-cancerous gastric tissues. However, NhTERT in samples of non-cancerous tissues was only approximately 14.8% of those found in the cancerous samples, and NhTERT was significantly higher (t = 12.76, P < 0.001) in tumor tissues (6.27 ± 0.89) than in the corresponding non-cancerous tissues (0.93 ± 0.18). We believed that these were not in contradiction to the findings using TRAP with which telomerase activity could not be detected in most non-malignant gastric tissues and the positive rate of hTERT mRNA expression by means of in situ hybridization or classical RT-PCR in gastric carcinoma[12-15]. The difference could be explained by the increased sensitivity of the RQ-PCR assay and the presence of residual malignant cells, inflammatory lymphocytes or tumor infiltrating activated cells. Our results showed real-time measurement of hTERT expression could discriminate gastric carcinoma from nonmalignant gastric tumors. This discrimination would be more distinct if the percentage of tumor cells was higher in the selected tissues.
No relationship was found in our study between hTERT mRNA expression level and patients’ age, gender, tumor size, location and tumor stage (pTNM) (P > 0.05) and there were statistical links between hTERT mRNA expression levels and the degree of differentiation (P = 0.007). These suggest that the up-regulation of hTERT appeared to be an early event in gastric carcinogenesis. HTERT may play a critical role in gastric carcinogenesis and have potential as a biomarker in the telomerase status for the early detection of gastric cancer. Real-time quantitative analysis of hTERT mRNA in tissues, cancer cells in the blood, and plasma or serum of the patients with cancer before and after surgery, radiotherapy or chemotherapy may be helpful for early detection and diagnosis of cancer, and find out therapeutic effect and estimate prognosis[16-21]. hTERT may become a target in gene therapy of cancer and inaugurate a new approach for cancer therapy.
In conclusion, monitoring hTERT mRNA expression with RQ-PCR analysis appears to be a new effective and sensitive method to better differentiate gastric cancer from nonmalignant gastric tumors. HTERT mRNA expression status may be used as a molecular marker of gastric cancer. These findings must be confirmed in a larger series of gastric cancer patients.
Edited by Chen WW and Wang XL Proofread by Xu FM
| 1. | Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 2004] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 2. | Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1302] [Cited by in RCA: 1318] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 3. | Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 892] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 4. | Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J. The RNA component of human telomerase. Science. 1995;269:1236-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1606] [Cited by in RCA: 1606] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 5. | Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1680] [Cited by in RCA: 1660] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 6. | Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2695] [Cited by in RCA: 2598] [Article Influence: 103.9] [Reference Citation Analysis (0)] |
| 7. | PE Biosystems. DNA/RNA real-time quantitative PCR.. Available from: http: //www.wzw.tum.de/gene-quantification/pe-realtimeoverview-1.pdf. |
| 8. | ABI Prism 7700 Sequence detection System User Bulletin #2(2001) Relative quantification of gene expression.. Available from: http: //docs.appliedbiosystems.com/pebiodocs/ 04303859.pdf. |
| 9. | Bièche I, Noguès C, Paradis V, Olivi M, Bedossa P, Lidereau R, Vidaud M. Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6:452-459. [PubMed] |
| 10. | Pfaffl MW, Hageleit M. Validities of mRNA quantification using recombinant RNA and recombinant DNA external cali-bration curves in real-time RT-PCR. Biotechnol Lett. 2001;23:275-282. [DOI] [Full Text] |
| 11. | Ke LD, Chen Z, Yung WK. A reliability test of standard-based quantitative PCR: exogenous vs endogenous standards. Mol Cell Probes. 2000;14:127-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 79] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Yoo J, Park SY, Kang SJ, Kim BK, Shim SI, Kang CS. Expression of telomerase activity, human telomerase RNA, and telomerase reverse transcriptase in gastric adenocarcinomas. Mod Pathol. 2003;16:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Miyachi K, Fujita M, Tanaka N, Sasaki K, Sunagawa M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J Exp Clin Cancer Res. 2002;21:269-275. [PubMed] |
| 14. | Okusa Y, Ichikura T, Mochizuki H, Shinomiya N. Clinical significance of telomerase activity in biopsy specimens of gastric cancer. J Clin Gastroenterol. 2000;30:61-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Yao XX, Yin L, Sun ZC. The expression of hTERT mRNA and cellular immunity in gastric cancer and precancerosis. World J Gastroenterol. 2002;8:586-590. [PubMed] |
| 16. | Dasí F, Lledó S, García-Granero E, Ripoll R, Marugán M, Tormo M, García-Conde J, Aliño SF. Real-time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA: a simple blood test to monitor disease in cancer patients. Lab Invest. 2001;81:767-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 71] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Buttitta F, Pellegrini C, Marchetti A, Gadducci A, Cosio S, Felicioni L, Barassi F, Salvatore S, Martella C, Coggi G. Human telomerase reverse transcriptase mRNA expression assessed by real-time reverse transcription polymerase chain reaction predicts chemosensitivity in patients with ovarian carcinoma. J Clin Oncol. 2003;21:1320-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Miura N, Shiota G, Nakagawa T, Maeda Y, Sano A, Marumoto A, Kishimoto Y, Murawaki Y, Hasegawa J. Sensitive detection of human telomerase reverse transcriptase mRNA in the serum of patients with hepatocellular carcinoma. Oncology. 2003;64:430-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | de Kok JB, Ruers TJ, van Muijen GN, van Bokhoven A, Willems HL, Swinkels DW. Real-time quantification of human telomerase reverse transcriptase mRNA in tumors and healthy tissues. Clin Chem. 2000;46:313-318. [PubMed] |
| 20. | Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823-3826. [PubMed] |
| 21. | Tchirkov A, Rolhion C, Kémény JL, Irthum B, Puget S, Khalil T, Chinot O, Kwiatkowski F, Périssel B, Vago P. Clinical implications of quantitative real-time RT-PCR analysis of hTERT gene expression in human gliomas. Br J Cancer. 2003;88:516-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |