Published online Aug 15, 2004. doi: 10.3748/wjg.v10.i16.2340
Revised: February 15, 2004
Accepted: February 24, 2004
Published online: August 15, 2004
AIM: To construct and select antigen epitopes of vacuolating cytotoxin A (VacA) for nontoxic VacA vaccine against Helicobacter pylori (H pylori ) infection.
METHODS: Eleven VacA epitopes were predicted according to VacA antigenic bioinformatics. Three candidates of VacA epitope were constructed through different combined epitopes. The candidate was linked with E. coli heat-labile enterotoxin B (LTB) by a linker of 7 amino acids, and cloned into plasmid pQE-60 in which fusion LTB-VacA epitope was efficiently expressed. To test the antigencity of the candidate, 6 BALB/c mice were treated with the fusion LTB-VacA epitope through intraperitoneal injection. To explore the ability of inhibiting the toxicity of VacA,cantiserum against the candidate was used to counteract VacA that induced HeLa cells to produce cell vacuoles in vitro.
RESULTS: Serum IgG against the candidate was induced in the BALB/c mice. In vitro, the three antisera against the candidate efficiently counteracted the toxicity of VacA, and decreased the number of cell vacuoles by 14.17%, 20.20% and 30.41% respectively.
CONCLUSION: Two of the three candidates, LZ-VacA1and LZ-VacA2, can be used to further study the mechanism of vacuolating toxicity of VacA, and to construct nontoxic VacA vaccine against H pylori infection.
-
Citation: Liu XL, Li SQ, Liu CJ, Tao HX, Zhang ZS. Antigen epitope of
Helicobacter pylori vacuolating cytotoxin A. World J Gastroenterol 2004; 10(16): 2340-2343 - URL: https://www.wjgnet.com/1007-9327/full/v10/i16/2340.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i16.2340
Pathogenic strains of Helicobacter pylori (H pylori) release a Mr 95000 protein toxin in the growth medium. Growing evidence indicates that VacA is a major virulence factor in H pylori long-term infection leading to gastroduodenal ulcers[1-5]. This toxin induces formation of vacuoles in the cytosol of cells, therefore it has been named vacuolating toxin[6-11]. VacA is thus considered as a therapeutic vaccine for individuals infected with H pylori. The VacA gene encodes a protoxin approximately Mr 140000, which belongs to the family of secreted proteins. During the process of VacA secretion, a Mr 95000 mature toxin is exported. A large oligomeric complex appear as ‘flower’ with Mr 900 000 which is composed of 6-7 VacA monomers[12-15]. When exposed to the acidic situation, the oligomeric complexes were assembled into monomers and the toxicity of VacA was enhanced[16-18]. After VacA exerted its effect on the cells for 90 min, vacuoles were formed[19]. The vacuoles induced by VacA were acidic. Intracellular vacuolation is believed to induce cell damage and eventually apoptosis, which might lead to release of necrotic factors in vivo and therefore contribute to the establishment of a chronic inflammatory response. Because vacuolating cytotoxin A is difficult to express, purify and construct the combined vaccine, we studied the antigen epitopes to reduce the toxin.
E. coli JM109, pFS2.2 and Hela cell were preserved in our laboratory; pQE-60 was a gift of professor Hou-Chu Zhu, Bejing Institute of Biotechnology, Beijing, China; H pylori Sydney strain (HP SS1), was a gift of professor Min-Hu Chen, Sun Yat-sen University, Guangzhou, China.
BamH I, EcoR V, Nco I, Hind III, Pyrobest Taq DNA polymerase and T4 DNA ligase were purchased from TaKaRa Biotechnology corporations.
Isopropyl β -D-thiogalactoside (IPTG), Freund’s adjuvant and sheep anti-mouse IgG-HRP were purchased from Sigma corporations, RPMI 1640 and newborn calf serum were purchased from Hyclone corporations.
BALB/c female mice: 6-8 wk old, SPF.
According to the protein characteristics of hydrophilicity, hydrophobicity, secondary structure, accessibility, flexibility and antigenicity, 9 VacA antigen epitopes in the amino acid site of 34-810 were predicted by the antigen-analyzing software GOLDKEY (Table 1).
| Site of amino acid | Amino acid sequences of predicted epitopes |
| 35-46 | AEEANKTPDKPD |
| 61-66 | PHKEYD |
| 146-154 | KDSADRTTR |
| 297-317 | GYKDKPKDKPSNTTQNNANNN |
| 335-338 | NSAQ |
| 446-450 | TDTKN |
| 566-568 | SGE |
| 734-737 | NNNR |
| 746-748 | TDD |
| 766-768 | DNY |
| 799-806 | TPTENGGN |
Three candidates of VacA antigen epitope were designed by combining part of 9 predicted epitopes. The candidate LZ-VacA1 was composed of amino acids 35-36 and 146-154, LZ-VacA2 included amino acids 297-317, and LZ-VacA3 contained amino acids 61-66, 446-450, 734-737, 746-748, 766-768, and 799-806. The gene sequence of candidate epitope was fused with a 7-amino acid linker (YPQDPSS). The nucleotide acid and amino acid sequences of three candidate epitopes were as following.
Two splicing sequences were designed according to the nucleotide acid sequences of the candidate epitope and linker. These sequences were synthesized artificially and spliced by PCR reaction. PCR reaction solution containing 5 μL 10 × PCR buffer, 0.25 μL pyrobest Taq polymerase (5 U/μL), 1 μL P1 and P2 primer (50 ng/μL), 4 μL dNTP, H2O to 50 μL. Five PCR cycles were performed, each at 95 °C for 30 s, at 60 °C for 30 s, at 72 °C for 15 s.
LZ-VacA1 primer sequence was P1: 5’--cag gat ccg tct tcc gcc gaa gaa gcc aat aaa acc cca gat aaa ccc gat aag--3’, P2: 5’---atc tct cgt ggt gcg atc agc act atc ctt atc ggg ttt atc tgg gg---3’.
LZ-VacA2 primer sequence was P1: 5’--cag gat ccg tct tcc ggt tat aag gat aaa cct aag gat aaa cct agt aac acc--3’, P2: 5’---atc gtt att att agc att att ttg cgt ggt gtt act agg ttt atc c---3’.
LZ-VacA3 primer sequence was, P1: 5’--cag gat ccg tct tcc cct cac aag gaa tac gac acg gat acc aaa aac aac aat aac cgc act gat--3’, P2: 5’---atc att gcc acc att ctc agt agg ggt gta att gtc gtc atc agt gcg gtt att gtt gt---3’.
The DNA sequences of the candidate epitope and plasmid pFS2.2 containing LTB, were digested with restriction endonucleases BamHI and EcoRV, and ligated with T4 ligase. The recombinant plasmid, which included LTB-VacA gene was transformed to JM109. The positive clones were screened and named them as pLZ-SV1, pLZ-SV2 and pLZ-SV3, respectively.
To improve the efficiency of expression, two primers with the cloning sites NcoI and HindIII were designed to construct plasmid pQE-60 with LTB-VacA gene. The recombinant plasmid was transformed to JM109. The positive clones were screened and named them as pLZ-QV1, pLZ-QV2 and pLZ-QV3 respectively.
The strain with recombinant plasmid was cultured in the Luria-Bertani broth for 3 h, induced by IPTG for 4 h, and then harvested. The targeted protein of LTB-VacA was an inclusion body by SDS-PAGE test. The inclusion body was denatured with 6 mol/L guanidine hydrochloride, and natured with dialysis. LTB-VacA infused protein was purified through the anti-LTB antibody affinity chromatography.
Twenty-four female BALB/c mice were randomly and averagely divided into control (LTB), LTB-VacA1, LTB-VacA2 and LTB-VacA3 groups. The mice of each group were immunized through intraperitoneal injection of 200 μL (100 μg protein) LTB, LTB-VacA1, LTB-VacA2 or LTB-VacA3 on days 0, 14 and 28. On days 7, 21 and 35, blood of each mouse was collected and antibody titer was determined.
VacA was purified from H pylori strain SS1 culture supernatant with ammonia sulfate precipitation. The preliminary experiment was performed to show the amount of H pylori strain SS1 culture supernatant was added when cell vacuoles were formed. HeLa cells were cultured as monolayers in flasks in RPMI 1640 containing NCS under 50 mL/L CO2 at 37 °C. Twenty-four hours before experiment, the cells were released with trypsin/EDTA and seeded in 96-well plates in 103/well. After the VacA protein was incubated with antibody to LTB-VacA1, 2, 3 and LTB for 4 h at 37 °C, we added the fixture and VacA protein onto the cell surface for 6 h. Then we calculated the total cell number and cell number of vacuolization.
Data are presented as mean ± SD. Analysis of variance with a two-tailed students t-test was used to identify significant differences. P < 0.05 was considered statistically significant.
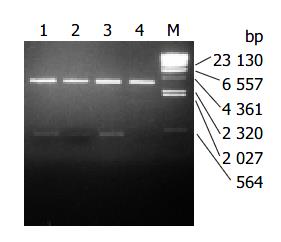
Recombinant plasmids pLZ-VacA1, 2, 3 encoded the infused gene of LTB and VacA1, 2, 3. It was shown from the digestion map of restriction endonucleases NcoI and Hind III, the infused gene was successfully cloned to PQE-60 (Figure 1). LTB-VacA1 had the nucleotide acid number of 465 bp (LTB387 + LZ-VacA1 78). LTB-VacA2 had the nucleotide acid number of 465 bp (LTB387 + LZ-VacA2 78). LTB-VacA3 had the nucleotide acid number of 489 bp (LTB387 + LZ-VacA3 102). The amino acids of three LTB-VacA were deduced from the nucleotide acid sequences. LTB-VacA1 and LTB-VacA2 had 155 amino acids, and the Mr was about 17000. LTB-VacA3 had 163 amino acids, and the molecular weight was about 17900. The sequences of the three genes were correct by sequencing analyses.
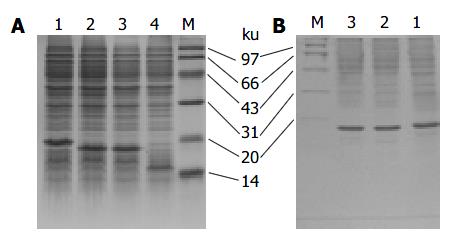
The LTB-VacA1, 2, 3 proteins were expressed in the JM109 strain. In the SDS-PAGE, the three proteins were 14.13%, 15.51% and 14.79% of the total protein respectively. After purification with anti-LTB antibody affinity chromatography, the percentage of three proteins in the total proteins was improved to 69.26%, 70.18% and 75.35% respectively (Figure 2).
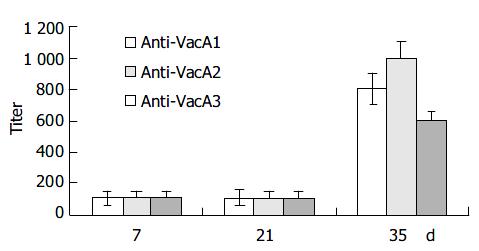
Intraperitoneal immunization with the infused protein and Freund’s adjuvant resulted in a marked elevation of serum IgG antibody in all the 6 mice after three immunizations (Figure 3).
Seven days after the first immunization, the Level of antibody was lower. The titer of the antibody was about 1:100. Seven days after the second immunization, the titer of the antibody had no remarkable changes. But 14 d after the third immunization, the titer of the anti-LTB-VacA1 antibody, the titer of the anti-LTB-VacA2 antibody and the titer of the anti-LTB-VacA1 antibody was increased to 1:800, 1:1000 and 1:600 respectively. The biggest value of positive and negative in the three antibodies was 3.8, 4.2 and 3.2.
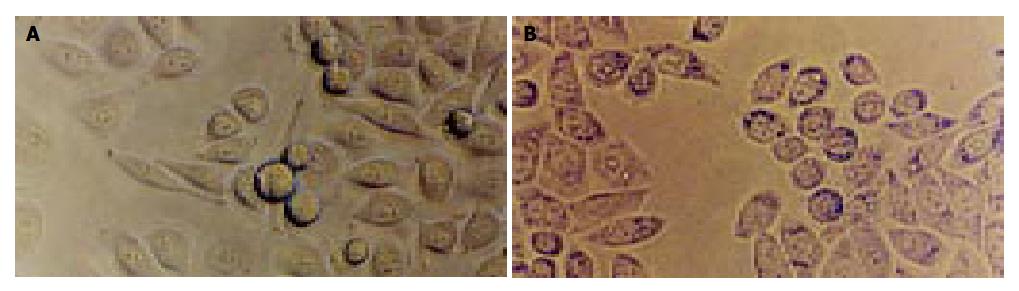
The preliminary experimental results showed when we added 10, 30, 40, 80 and 100 μL culture supernatant, the cell vacuolization was produced. Because the space of wells in flasks well was limited, 100 μL VacA protein was added into the culture medium. The ratio of cell vacuolization after we added 100 μL protein was 49.52% (Figure 4).
The control serum, anti-VacA1 serum, anti-VacA2 serum and anti-VacA1 serum were incubated with 100 μL VacA antigen for 4 h. Then we added the fixture and VacA protein in the flasks wells for 6 h. The result showed the control serum and 100 μL VacA antigen could not reduce the ratio of cell vacuolization, but anti-VacA1 serum, anti-VacA2 serum, anti-VacA3 serum could decrease the ratio of cell vacuolization. The change rate of cell vacuolization is 30.41%, 20.20% and 14.17% respectively (Table 2).
Many methods could predict the epitopes of protein known as the primary structure, for example hydrophilicity scheme[20], accessibility scheme[21], antigenicity scheme[22], flexibility scheme[23,24] and secondary structure scheme[17]. The antigenic epitopes are correlated with the characteristics, number, sequence of amino acid and protein conformation. Because the different prediction methods emphasize different biological information of antigens, several methods are considered in practice. In this study we chose the GOLDKEY software developed by the Experimental Group led by professor Jia-Jin Wu to analyse the characteristics of VacA protein including hydrophilicity, hydrophobicity, secondary structure, accessibility, flexibility and antigenicity. This software could predict the liner B epitope. At last we got 11 VacA candidate epitopes.
Due to the small molecular weight and the weakness of antigenicity of the antigen epitopes, the carrier or adjuvant must be linked to the epitopes[25,26]. Several epitopes were joined in series and at last got 3 VacA candidate epitopes. LTB is an excellent protein adjuvant to facilitate the organism to produce the antibody epitopes, so we chose LTB to link the 3 epitopes. The experiment of Schodel enucleate that plasmid pFS2.2 was a carrier in which LTB gene could express soluble LTB and carry outer polypeptides. The experiment of Zhang showed that 21-bp nucleotide acids between LTB and the epitopes could enhance the antigencity of the epitopes. In our study, 7 peptides were used as a linker to join LTB with the epitopes.
At the beginning, LTB-VacA was cloned into plasmid pFS2.2, but the gene could not express these proteins, and then the genes were cloned into plasmid pET22b ( + ) again. There was a signal peptide in plasmid pET22b ( + ) in which the gene could express the soluble protein and secrete the protein into periplasm. The soluble proteins would be purified through the anti-LTB antibody affinity chromatography. Contrary to our wishes, the proteins in pFS2.2 were not expressed as expected. Finally, infused proteins were cloned into plasmid pQE-60 and expressed in JM109, but the expressed proteins were inclusion bodies. The inclusion bodies were denatured with guanidine hydrochloride and natured with dialysis, and LTB-VacA was purified through the anti-LTB antibody affinity chromatography.
Protein vacuolating toxin A is the only known virulence factor of H pylori. Ninety minutes after VacA activation, the acidic vacuoles were induced in cells. Scientific researches showed that VacA was integrated with receptors in membranes to form an anionic channel. This channel could change the permeation characteristics of the membranes, so that the cells were damaged would undergo apoptosis. In this study, all the 3 antigen epitopes of VacA could induce antibody in mice. Although the antibody could inhibit the vacuolation of Hela cells at a certain extent, they did not inhibit the vacuoles entirely. There are two reasons for this result. First, these epitopes were a part of the neutralized epitopes of VacA, antibody to these epitopes combined with VacA did not destroy the toxicity of VacA, only suppressed the toxin partly. Second, the titer of the antibody was not enough to neutralize the toxin of VacA. Next we are going to settle the problem. First, these epitopes will be joined in series for several copies so that the titer of the antibody will be improved to inhibit the toxin of VacA. Second, we will construct deficient mutations to farther verify the neutralized epitopes of VacA.
Edited by Wang XL and Chen WW Proofread by Xu FM
| 1. | Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653-1658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 435] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 2. | van Amsterdam K, van Vliet AH, Kusters JG, Feller M, Dankert J, van der Ende A. Induced Helicobacter pylori vacuolating cytotoxin VacA expression after initial colonisation of human gastric epithelial cells. FEMS Immunol Med Microbiol. 2003;39:251-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Inui T, Mizuno S, Takai K, Nakagawa M, Uchida M, Fujimiya M, Asakawa A, Inui A. Helicobacter pylori cytotoxin: a novel ligand for receptor-like protein tyrosine phosphatase beta (review). Int J Mol Med. 2003;12:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 4. | Cho SJ, Kang NS, Park SY, Kim BO, Rhee DK, Pyo S. Induction of apoptosis and expression of apoptosis related genes in human epithelial carcinoma cells by Helicobacter pylori VacA toxin. Toxicon. 2003;42:601-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Toro Rueda C, García-Samaniego J, Casado Fariñas I, Rubio Alonso M, Baquero Mochales M. [Clinical importance of the CagA and VacA proteins and of the host factores in the development of peptic ulcer in patients infected by Helicobacter pylori]. Rev Clin Esp. 2003;203:430-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 6. | Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, Jellum E, Orentreich N, Vogelman JH, Friedman GD. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1287] [Cited by in RCA: 1229] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 7. | Yuan JP, Li T, Shi XD, Hu BY, Yang GZ, Tong SQ, Guo XK. Deletion of Helicobacter pylori vacuolating cytotoxin gene by introduction of directed mutagenesis. World J Gastroenterol. 2003;9:2251-2257. [PubMed] |
| 8. | Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 409] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 9. | Qiao W, Hu JL, Xiao B, Wu KC, Peng DR, Atherton JC, Xue H. cagA and vacA genotype of Helicobacter pylori associated with gastric diseases in Xi'an area. World J Gastroenterol. 2003;9:1762-1766. [PubMed] |
| 10. | Caputo R, Tuccillo C, Manzo BA, Zarrilli R, Tortora G, Blanco Cdel V, Ricci V, Ciardiello F, Romano M. Helicobacter pylori VacA toxin up-regulates vascular endothelial growth factor expression in MKN 28 gastric cells through an epidermal growth factor receptor-, cyclooxygenase-2-dependent mechanism. Clin Cancer Res. 2003;9:2015-2021. [PubMed] |
| 11. | Wang J, van Doorn LJ, Robinson PA, Ji X, Wang D, Wang Y, Ge L, Telford JL, Crabtree JE. Regional variation among vacA alleles of Helicobacter pylori in China. J Clin Microbiol. 2003;41:1942-1945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570-10575. [PubMed] |
| 13. | Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801-807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 134] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Nguyen VQ, Caprioli RM, Cover TL. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun. 2001;69:543-546. [PubMed] |
| 15. | Lanzavecchia S, Bellon PL, Lupetti P, Dallai R, Rappuoli R, Telford JL. Three-dimensional reconstruction of metal replicas of the Helicobacter pylori vacuolating cytotoxin. J Struct Biol. 1998;121:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Cover TL, Hanson PI, Heuser JE. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 162] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 17. | Yahiro K, Niidome T, Kimura M, Hatakeyama T, Aoyagi H, Kurazono H, Imagawa Ki, Wada A, Moss J, Hirayama T. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J Biol Chem. 1999;274:36693-36699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 129] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | McClain MS, Cao P, Cover TL. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect Immun. 2001;69:1181-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Cover TL, Halter SA, Blaser MJ. Characterization of HeLa cell vacuoles induced by Helicobacter pylori broth culture supernatant. Hum Pathol. 1992;23:1004-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981;78:3824-3828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2408] [Cited by in RCA: 2505] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 21. | Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1590] [Cited by in RCA: 1533] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 22. | Welling GW, Weijer WJ, van der Zee R, Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS Lett. 1985;188:215-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 237] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 23. | Zhao S, Goodsell DS, Olson AJ. Analysis of a data set of paired uncomplexed protein structures: new metrics for side-chain flexibility and model evaluation. Proteins. 2001;43:271-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Kolibal SS, Brady C, Cohen SA. Definition of epitopes for monoclonal antibodies developed against purified sodium channel protein: implications for channel structure. J Membr Biol. 1998;165:91-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |