Published online Aug 15, 2004. doi: 10.3748/wjg.v10.i16.2318
Revised: June 22, 2003
Accepted: July 30, 2003
Published online: August 15, 2004
AIM: In order to elucidate the molecular mechanism of lymphatic metastasis of hepatocarcinoma, we detected the difference of gene expression between mouse hepatocarcinoma cell lines Hca-F and Hca-P with different lymphatic metastasis potential.
METHODS: cDNA of Hca-F cells was used as a tester and cDNA of Hca-P cells was used as a driver. cDNAs highly expressed in Hca-F cells were isolated by the suppression subtractive hybridization (SSH) method. The isolated cDNA was cloned into T/A cloning vector. The ligation products were transformed into DH5 α competent cells. Individual clones were randomly selected and used for PCR amplification. Vector DNA from positive clones was isolated for sequencing.
RESULTS: There were 800 positive clones in amplified subtracted cDNA library. Random analysis of 160 clones with PCR showed that 95% of the clones contained 100-700 bp inserts. Analysis of 20 sequenced cDNA clones randomly picked from the SSH library revealed 4 known genes (mouse heat shock protein 84 ku, DNA helicase, ribosomal protein S13 ,ethanol induced 6 gene) and 3 expressed sequence tags (ESTs). Four cDNAs showed no homology and presumably represent novel genes.
CONCLUSION: A subtracted cDNA library of differentially expressed genes in mouse heptocarcinoma cell lines with different lymphatic metastasis potential was successfully constructed with SSH and T/A cloning techniques. The library is efficient and lays a solid foundation for searching new lymphatic metastasis related genes. The expression of mouse heat shock protein gene, DNA helicase and other 4 novel gene may be different between mouse heptocarcinoma cell lines with different lymphatic metastasis potential.
- Citation: Hou L, Tang JW, Cui XN, Wang B, Song B, Sun L. Construction and selection of subtracted cDNA library of mouse hepatocarcinoma cell lines with different lymphatic metastasis potential. World J Gastroenterol 2004; 10(16): 2318-2322
- URL: https://www.wjgnet.com/1007-9327/full/v10/i16/2318.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i16.2318
Metastasis is the most lethal attribute of malignant tumors[1]. Ninety percent of malignant tumors are carcinomas, and lymph nodes are often the first organ to develop metastasis[2]. Lymph node metastases form a bridgehead for further metastatic spread. But its molecular mechanism remains poorly understood. A mouse hepatocarcinoma cell line named Hca-F with high lymphogenous metastatic potential and its syngeneic cell line named Hca-P[3] with low lymphogenous metastatic potential have been isolated from hepatocarcinomas in mice. The biological phenotypes between different cells are based on the difference between their phenotypes of gene expression. In this study, we constructed a subtracted cDNA library of differentially expressed genes in these two hepatocarcinoma cell lines using the suppressive subtractive hybridization (SSH)[4-6], which can detect the difference in gene expression between different cells.
Hepatocarcinoma cell lines, Hca-F and Hca-P, were obtained from our department. Twenty inbred 615-mice maintained in our laboratory were equally divided into 2 groups. Hca-F and Hca-P cells (2 × 10 6 cells/mouse) were respectively inoculated in 10 mice in each group. On the 28th day post inoculation, the mice were killed and their lymph nodes were collected and stained by HE and examined under microscope. Therefore, the lymph node metastasis rate was calculated.
Total RNA was isolated from Hca-F and Hca-P cells (108 cells) respectively using Trizol regent (Gibco BRL, USA). And mRNA was isolated from total RNA using the OligotexTM mRNA kit (Qiagen, USA) according to its manufacturer’s instructions. The integrity of RNA and mRNA was checked on a 1% agrose gel.
Analysis of differentially expressed genes in Hca-F and Hca-P cells was performed by the suppression subtractive hybridization (SSH) using the CLONTECH PCR-SelectTM cDNA subtraction kit (Clotech, USA). In brief[7,8], 2 μg of mRNA from Hca-F and Hca-P cells was used for double strand cDNA synthesis, and the resulting cDNA was digested with Rsa I. The digested cDNA of Hca-F cells(as a Tester) was split into two groups and ligated to either adaptor I or adaptor 2R. Subtractive hybridization was performed by annealing an excess of Hca-P cDNAs (as a Driver) with each sample of adaptor ligated tester cDNAs. The cDNAs were heat denatured and incubated at 68 °Cfor 8 h. After the first hybridization, the two samples were mixed together and hybridized again with freshly denatured driver cDNAs for 20 h at 68 °C. The two rounds of hybridization would generate a normalized population of tester specific cDNA with different adaptors on each end. After filling in the end, two rounds of PCR amplification were performed to enrich desired cDNAs containing both adaptors by exponential amplification of these products. The optimized cycles for the first and second PCR were 30 and 20, respectively, to increase representation and reduce redundancy of subtracted cDNA libraries. Secondary PCR products were used as templates for PCR amplification of G3PDH at 20, 25, 30, 35 cycles to assure subtracted efficiency.
Products from the secondary PCR were T-A cloned into a T vector (Takara, China). The ligation products were transformed into DH5 α competent cells. The transformed cells were plated on LB plates containing ampicilin, X-Gal and IPTG, which allowed for color selection of colonies. Randomly selected individual white clones were grown for 8 h and then used for PCR amplification of clones.
Twenty candidate positive clones from subtracted cDNA library were selected for sequencing. Sequencing was performed by Takara Biotech using a M13 primer. The BLAST program was used to search for the cDNA sequence homology of isolated clones in GeneBank.
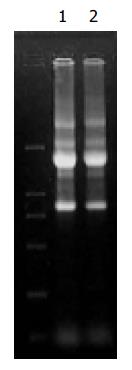
The A260/A280 of RNA and mRNA from Hca-F and Hca-P was 1.9, which showed extracted RNA and mRNA were pure. Agarose gel electrophoresis showed that RNA was not destroyed by ribonucleases (Figure 1).
The size of digested cDNA molecular was smaller than undigested in agarose gel electrophoresis, indicating that tester cDNA and driver cDNA were completely digested.
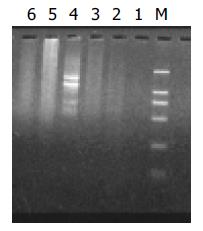
In this study, SSH was performed to identify differentially expressed genes among cDNAs of hepatocarcinoma cell lines, Hca-F and Hca-P. One subtracted F-P cDNA library was constructed using Hca-F with high lymphogenous metastatic potential as a tester and its syngeneic cell line named Hca-P with low lymphogenous metastatic potential as a driver. The subtracted cDNA library after secondary PCR amplification looked like smears (Figure 2).
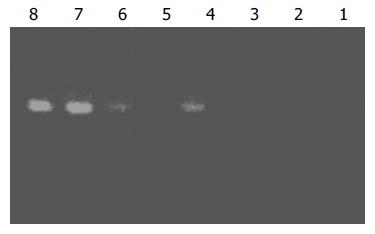
Subtraction efficiency analysis showed that the abundance of non-differentially expressed genes was effectively reduced. In no subtracted cDNA library, the PCR products of housekeeping gene G3PDH were visible after 25 cycles. However, 35 cycles were required in the subtracted cDNA library for G3PDH to be detected (Figure 3).
The subtraction cDNA library contained 800 positive (white) clones. Random analysis of 160 clones with PCR showed that 95% of positive clones contained 100-700 bp inserts (Figure 4).
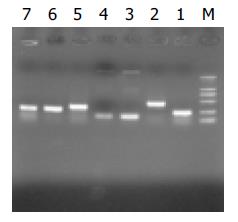
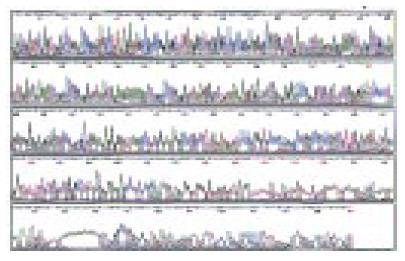
Automatic sequencing proved that most clones were isolated no more than two times (Figure 5, Figure 6, Figure 7). BLAST homology search revealed that some sequences were homologous with known gene fragments and others were possibly novel genes, showing few sequence homologies with any known sequences in the GenBank. The results of homology search are shown in Table 1.
| Clone number | Size(bp) | Sequence identity |
| 10-3 | 508 | Mus musculus heat shock protein, 84 ku 1 (Hsp84-1) |
| 2-2 | 385 | Mus musculus ribosomal protein S13 (Rps13) |
| 6-2 | 323 | Mouse mRNA for DNA helicase |
| 18-4 | 226 | Mouse, similar to ethanol induced 6, clone MGG51351 |
| 11-7 | 363 | Mouse 10 d embryo whole body cDNA, RIKEN full-length enrich library, clone 2610102k11 |
| 16-3 | 403 | Mouse 2 d neonate thymus thymic cells cDNA RIKEN full-length library enriched library, clone E430002F13 |
| 7-1 | 570 | Mouse chromosome 2 clone RP 24-322M5 |
| 15-7 | 460 | Mouse chromosome 12 clone RP23-210N16 |
| 1-8 | 200 | EST-mouse |
| 5-8 | 159 | EST-mouse |
| 6-8 | 216 | EST-mouse |
| 2-6 | 61 | Unknown |
| 1-3 | 132 | Unknown |
| 1-6 | 241 | Unknown |
| 16-8 | 240 | Unknown |
The lymphatic metastasis rate was 80% (8/10) and 10% (1/10), respectively, for Hca-F cells and Hca-P cells.
Cancer is the most important cause of death of humans. There is no technique that can selectively kill cancer cells. A wide range of Metastasis leads to the failure of treatment and the death of patients. Therefore, investigation of cancer metastasis, particular the mechanisms, is important in order to improve the efficacy of the treatment. Metastasis[9] is a complex process, which is made up of several steps. However, the molecular mechanism of lymphatic metastasis remains poorly understood because of lack of lymphatic metastasis models. A mouse hepatocarcinoma cell line named Hca-F with a metastasis rate over 70% and its syngeneic cell line named Hca-P with a metastasis rate less than 30% have been separated from hepatocacinomas in mice. The difference in their metastasis potential might be based on the difference in their phenotypes of gene expression. Many different techniques could be used to isolate differentially expressed genes, such as expressed sequencing tag (EST)[10], serial analysis of gene expression (SAGE)[11], subtractive hybridization, mRNA differential display (DD-RTPCR)[12], cDNA representative difference analysis (RDA)[13], suppression subtractive hybridization (SSH), and cDNA microarry[14].
The novel technique named SSH[15-18] has become an ideal subtractive system that combines high subtraction efficiency with normalized representation of differentially expressed genes, and is based on suppression PCR that permits exponential amplification of genes differing in abundance, but suppresses the amplification of sequences of identical abundance genes in two cells at the same time. Therefore, differentially expressed genes of low abundance can be cloned, while differentially expressed genes of high abundance are not excessively isolated. It has been reported that SSH can achieve more than 1000-fold enrichment of low abundance genes, but only requires 2 μg mRNA instead of the 5-20 μg required in other methods. This method has been successfully used to isolate significant genes in many researches[19-23]. Von Stein et al[24] observed a 94% positive rate in their study, and suggested that confirmation of differential expression by Northern blot analysis for each clone obtained was unnecessary.
In the present study, we reported the efficiency of SSH technique in identifying differentially expressed genes in Hca-F and Hca-P. PCR analysis showed that the subtraction efficiency after subtractive hybridization was effectively reduced. We constructed a subtracted cDNA library between Hca-F cells and Hca-P cells. There were 800 positive clones in the amplified subtracted cDNA library. Random analysis of 160 clones with PCR showed that 95% of clones contained 100-700 bp inserts. After sequencing and BLAST homology search, these clones could be divided into three groups: known genes preciously reported to be related to metastasis and carcinogenesis, known genes never described to be related to metastasis and carcinogenesis, and unknown genes. The identification of the first group of known genes attested to the validity of this method. The library is efficient and lays a solid foundation for searching new lymphatic metastasis related genes.
The No. 10-3 clone obtained in the present study showed homology with mouse heat shock proteins(HSPs). HSPs[25-27] have been found to be ubiquitous molecules expressed in response to stress in all living organisms. The three important roles[28] in regard to cancer development are regulation of apoptosis, modulation of immune response and drug resistance. HSPs are cytoplasmic proteins that could act as molecular chaperones for protein molecules in various intra-cellular processes[29,30]. They are called “heat shock proteins” since they were first discovered in cells exposed to high temperatures. HSPs were also expressed in hepatocarcinoma (HCC)[31-34], and up-regulated in early stage of HCC compared with noncancerous liver tissue. King et al[33] reported that HSPs expressed in HCC (45/58), and its expression was stronger in HCC than in noncancerous liver tissue. HSPs were related to metastasis[35-38]. HSP expression was found in 54 out of 86 (62.7%) gastric carcinomas and was significantly related to more than six metastatic lymph nodes[35]. Over expression of HSPs was related to tumor configuration, lymph node metastasis, and lymphatic vessel invasion in esophageal squamous cell carcinoma[36]. So far, there has no report concerning HSP expression and lymphatic metastasis of HCC. In the present study, we found that HSP expression was different between mouse HCC cell lines Hca-F and Hca-P, indicating that HSP expression might be related to HCC metastasis.
The No. 6-2 clone obtained showed homology with mouse helicase. It has been found that DNA helicases[39-41], because they unwound duplex DNA, have important roles in cellular DNA events such as replication, recombination, repair and transcription. The change in helicases expression could be found in human carcinomas[42], lymphoma[43], leukemia and so on. Helicases were also related to metastasis[44]. B16-F10 and B16-BL6 are B16 mouse melanoma sublines that preferentially metastasize to the lung and their metastasis potential is different. Helicases and ribosomal protein (L37) showed higher expression in B16-BL6 cells than in B16-F10 cells[45]. There has been no report on the association between helicase up-regulation and HCC metastasis. Thus, further investigations are required to elucidate the role of helicase in HCC metastasis.
Besides, two clones obtained in the present study showed homology to mouse ribosomal protein S13 and ethanol induced 6 gene. To our knowledge, there have been no reports on their relation to metastasis. Whether ribosomal protein S13 and ethanol induced 6 gene can be considered as candidate gene of metastasis related gene remains unclear. Clone 11-7 showed homology with mouse of 10-d embryo whole body cDNA library clone 2610102k11, It is important to study the correlation between metastasis and embryo gene expression. Clone 7-1 showed homology with mouse mouse chromosome 2 clone RP 24-322M5 and clone 15-7 showed homology with mouse chromosome 12 clones RP23-210N16. Whether there are candidate metastasis-related genes on chromosome 2 and chromosome 12 needs further investigation. The other three clones showed homology with three EST sequence.
In addition to the known genes, four clones showed no homology with any known sequences, indicating that they were novel genes, although the full-length sequence of these new genes and their function in lymphatic metastasis remain to be determined.
In conclusion, differentially expressed genes in mouse hepatocarcinoma cell lines with different lymphatic metastasis potential can be isolated by SSH method. Comprehensive study of these genes can help understand the molecular mechanism of lymphatic metastasis.
Edited by Xia HHX and Wang XL Proofread by Chen WW and Xu FM
| 1. | Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst. 2000;92:1717-1730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 182] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 2. | Sleeman JP. The lymph node as a bridgehead in the metastatic dissemination of tumors. Recent Results Cancer Res. 2000;157:55-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 3. | Hou L, Li Y, Jia YH, Wang B, Xin Y, Ling MY, Lü S. Molecular mechanism about lymphogenous metastasis of hepatocarcinoma cells in mice. World J Gastroenterol. 2001;7:532-536. [PubMed] |
| 4. | Ahmed FE. Molecular techniques for studying gene expression in carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2002;20:77-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Li J, Han BL, Huang GJ, Qian GS, Liang P, Yang TH, Chen J. [Screening and identification for cDNA of differentially expressed genes in human primary hepatocellular carcinoma]. Zhonghua Yixue Yichuanxue Zazhi. 2003;20:49-52. [PubMed] |
| 6. | Petersen S, Petersen I. Expression profiling of lung cancer based on suppression subtraction hybridization (SSH). Methods Mol Med. 2003;75:189-207. [PubMed] |
| 7. | Luo M, Kong XY, Liu Y, Zhou RH, Jia JZ. [cDNA libraries construction and screening in gene expression profiling of disease resistance in wheat]. Yichuan Xuebao. 2002;29:814-819. [PubMed] |
| 8. | Liu Y, Cheng J, Lu YY, Wang G, Mou JS, Li L, Zhang LX, Chen JM. [Cloning of genes transactivated by hepatitis B virus X protein]. Zhonghua Ganzangbing Zazhi. 2003;11:5-7. [PubMed] |
| 9. | Poste G, Fidler IJ. The pathogenesis of cancer metastasis. Nature. 1980;283:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1079] [Cited by in RCA: 1019] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 10. | Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1276] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 11. | Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2999] [Cited by in RCA: 2566] [Article Influence: 85.5] [Reference Citation Analysis (0)] |
| 12. | Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3487] [Cited by in RCA: 3168] [Article Influence: 96.0] [Reference Citation Analysis (0)] |
| 13. | Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 866] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 14. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5102] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 15. | Ai JK, Huang X, Wang YJ, Bai Y, Lu YQ, Ye XJ, Xin DQ, Na YQ, Zhang ZW, Guo YL. [Screening of novel genes differentially expressed in human renal cell carcinoma by suppression subtractive hybridization]. Ai Zheng. 2002;21:1065-1069. [PubMed] |
| 16. | Li JY, Boado RJ, Pardridge WM. Blood-brain barrier genomics. J Cereb Blood Flow Metab. 2001;21:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 108] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Chen I, Hsieh T, Thomas T, Safe S. Identification of estrogen-induced genes downregulated by AhR agonists in MCF-7 breast cancer cells using suppression subtractive hybridization. Gene. 2001;262:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Wang H, Zhan Y, Xu L, Feuerstein GZ, Wang X. Use of suppression subtractive hybridization for differential gene expression in stroke: discovery of CD44 gene expression and localization in permanent focal stroke in rats. Stroke. 2001;32:1020-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Zhou J, Wang H, Lu A, Hu G, Luo A, Ding F, Zhang J, Wang X, Wu M, Liu Z. A novel gene, NMES1, downregulated in human esophageal squamous cell carcinoma. Int J Cancer. 2002;101:311-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Saito A, Fujii G, Sato Y, Gotoh M, Sakamoto M, Toda G, Hirohashi S. Detection of genes expressed in primary colon cancers by in situ hybridisation: overexpression of RACK 1. Mol Pathol. 2002;55:34-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Miyasaka Y, Enomoto N, Nagayama K, Izumi N, Marumo F, Watanabe M, Sato C. Analysis of differentially expressed genes in human hepatocellular carcinoma using suppression subtractive hybridization. Br J Cancer. 2001;85:228-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Roberts D, Williams SJ, Cvetkovic D, Weinstein JK, Godwin AK, Johnson SW, Hamilton TC. Decreased expression of retinol-binding proteins is associated with malignant transformation of the ovarian surface epithelium. DNA Cell Biol. 2002;21:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Stassar MJ, Devitt G, Brosius M, Rinnab L, Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A, Zöller M. Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer. 2001;85:1372-1382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 117] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | von Stein OD, Thies WG, Hofmann M. A high throughput screening for rarely transcribed differentially expressed genes. Nucleic Acids Res. 1997;25:2598-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 117] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Piura B, Rabinovich A, Yavelsky V, Wolfson M. [Heat shock proteins and malignancies of the female genital tract]. Harefuah. 2002;141:969-72, 1010, 1009. [PubMed] |
| 26. | Friedman EJ. Immune modulation by ionizing radiation and its implications for cancer immunotherapy. Curr Pharm Des. 2002;8:1765-1780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 151] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 27. | Parmiani G, Castelli C, Dalerba P, Mortarini R, Rivoltini L, Marincola FM, Anichini A. Cancer immunotherapy with peptide-based vaccines: what have we achieved? Where are we going? J Natl Cancer Inst. 2002;94:805-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 283] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 28. | Lebret T, Watson RW, Fitzpatrick JM. Heat shock proteins: their role in urological tumors. J Urol. 2003;169:338-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 781] [Cited by in RCA: 743] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 30. | Candido EP. The small heat shock proteins of the nematode Caenorhabditis elegans: structure, regulation and biology. Prog Mol Subcell Biol. 2002;28:61-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, Hirohashi S. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 221] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Yin Y, Qin Q, Zhang W, Zhao J, Zhang C, Yu J. [Overexpression of heat shock protein 70 and spontaneous cancer cell apoptosis in hepatocellular carcinoma]. Zhonghua Ganzangbing Zazhi. 2001;9:84-85. [PubMed] |
| 33. | King KL, Li AF, Chau GY, Chi CW, Wu CW, Huang CL, Lui WY. Prognostic significance of heat shock protein-27 expression in hepatocellular carcinoma and its relation to histologic grading and survival. Cancer. 2000;88:2464-2470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Osada T, Sakamoto M, Nishibori H, Iwaya K, Matsuno Y, Muto T, Hirohashi S. Increased ubiquitin immunoreactivity in hepatocellular carcinomas and precancerous lesions of the liver. J Hepatol. 1997;26:1266-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Kapranos N, Kominea A, Konstantinopoulos PA, Savva S, Artelaris S, Vandoros G, Sotiropoulou-Bonikou G, Papavassiliou AG. Expression of the 27-kDa heat shock protein (HSP27) in gastric carcinomas and adjacent normal, metaplastic, and dysplastic gastric mucosa, and its prognostic significance. J Cancer Res Clin Oncol. 2002;128:426-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 36. | Noguchi T, Takeno S, Shibata T, Uchida Y, Yokoyama S, Müller W. Expression of heat shock protein 70 in grossly resected esophageal squamous cell carcinoma. Ann Thorac Surg. 2002;74:222-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Sagol O, Tuna B, Coker A, Karademir S, Obuz F, Astarcioglu H, Küpelioglu A, Astarcioglu I, Topalak O. Immunohistochemical detection of pS2 protein and heat shock protein-70 in pancreatic adenocarcinomas. Relationship with disease extent and patient survival. Pathol Res Pract. 2002;198:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | Mese H, Sasaki A, Nakayama S, Yoshioka N, Yoshihama Y, Kishimoto K, Matsumura T. Prognostic significance of heat shock protein 27 (HSP27) in patients with oral squamous cell carcinoma. Oncol Rep. 2002;9:341-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 39. | Ishiguro H, Shimokawa T, Tsunoda T, Tanaka T, Fujii Y, Nakamura Y, Furukawa Y. Isolation of HELAD1, a novel human helicase gene up-regulated in colorectal carcinomas. Oncogene. 2002;21:6387-6394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Fuchsová B, Novák P, Kafková J, Hozák P. Nuclear DNA helicase II is recruited to IFN-alpha-activated transcription sites at PML nuclear bodies. J Cell Biol. 2002;158:463-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Furuichi Y. Premature aging and predisposition to cancers caused by mutations in RecQ family helicases. Ann N Y Acad Sci. 2001;928:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 42. | Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum Mol Genet. 2001;10:741-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 157] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 43. | Kaneko H, Morimoto W, Fukao T, Kasahara K, Kondo N. Telomerase activity in cell lines and lymphoma originating from Bloom syndrome. Leuk Lymphoma. 2001;42:757-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 44. | Takagi Y, Suyama E, Kawasaki H, Miyagishi M, Taira K. Mechanism of action of hammerhead ribozymes and their applications in vivo: rapid identification of functional genes in the post-genome era by novel hybrid ribozyme libraries. Biochem Soc Trans. 2002;30:1145-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 45. | Ishiguro T, Nakajima M, Naito M, Muto T, Tsuruo T. Identification of genes differentially expressed in B16 murine melanoma sublines with different metastatic potentials. Cancer Res. 1996;56:875-879. [PubMed] |