Copyright
©The Author(s) 2021.
World J Gastroenterol. Jul 7, 2021; 27(25): 3790-3801
Published online Jul 7, 2021. doi: 10.3748/wjg.v27.i25.3790
Published online Jul 7, 2021. doi: 10.3748/wjg.v27.i25.3790
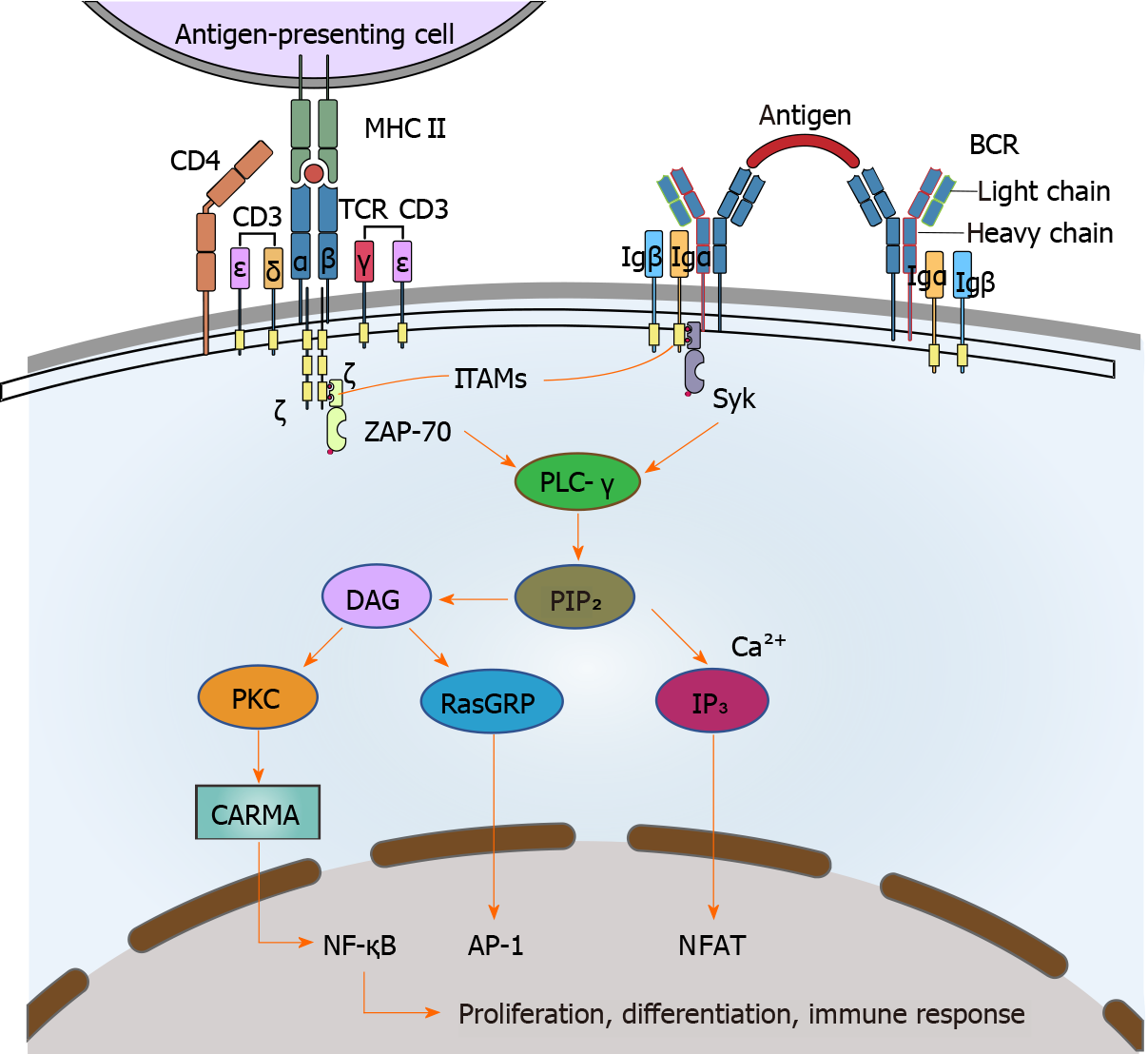
Figure 1 T-cell receptor and B-cell receptor structure and signaling pathways.
T-cell receptor and B-cell receptor complexes include both variable antigen-recognition proteins and invariant signaling proteins. Phosphorylation of the ITAMs in CD3 ε, γ, δ, and the ζ chain enables them to bind the cytosolic tyrosine kinase ZAP-70, which in turn recruits and activates PLC-γ. Activated PLC-γ cleaves PIP2 to yield DAG and IP3. IP3 increases intracellular Ca2+ concentration, activating calcineurin, a phosphatase that then activates an NFAT transcription factor. DAG recruits PKC to activate CARMA, which leads to activation of NF-κB and recruits RasGRP, which activates AP-1. These three important signaling pathways activate transcription factors in the nucleus, including NF-κB, NFAT, and AP-1, which result in cell differentiation, proliferation, and immune response. AP-1: Activator protein-1; CARMA: Caspase recruitment domain family, member 14 protein; DAG: Diacylglycerol; IP3: Inositol trisphosphate; ITAM: Immunoreceptor tyrosine-based activation motif; NFAT: Nuclear factor of activated T cells; NF-κB: Nuclear factor kappa B; PIP2: Phosphatidylinositol bisphosphate; PKC: Protein kinase C; PLC-γ: Phospholipase C-γ; RasGRP: RAS guanyl releasing protein; Syk: Spleen-associated tyrosine kinase; ZAP-70: Zeta chain of T-cell receptor-associated protein kinase 70.
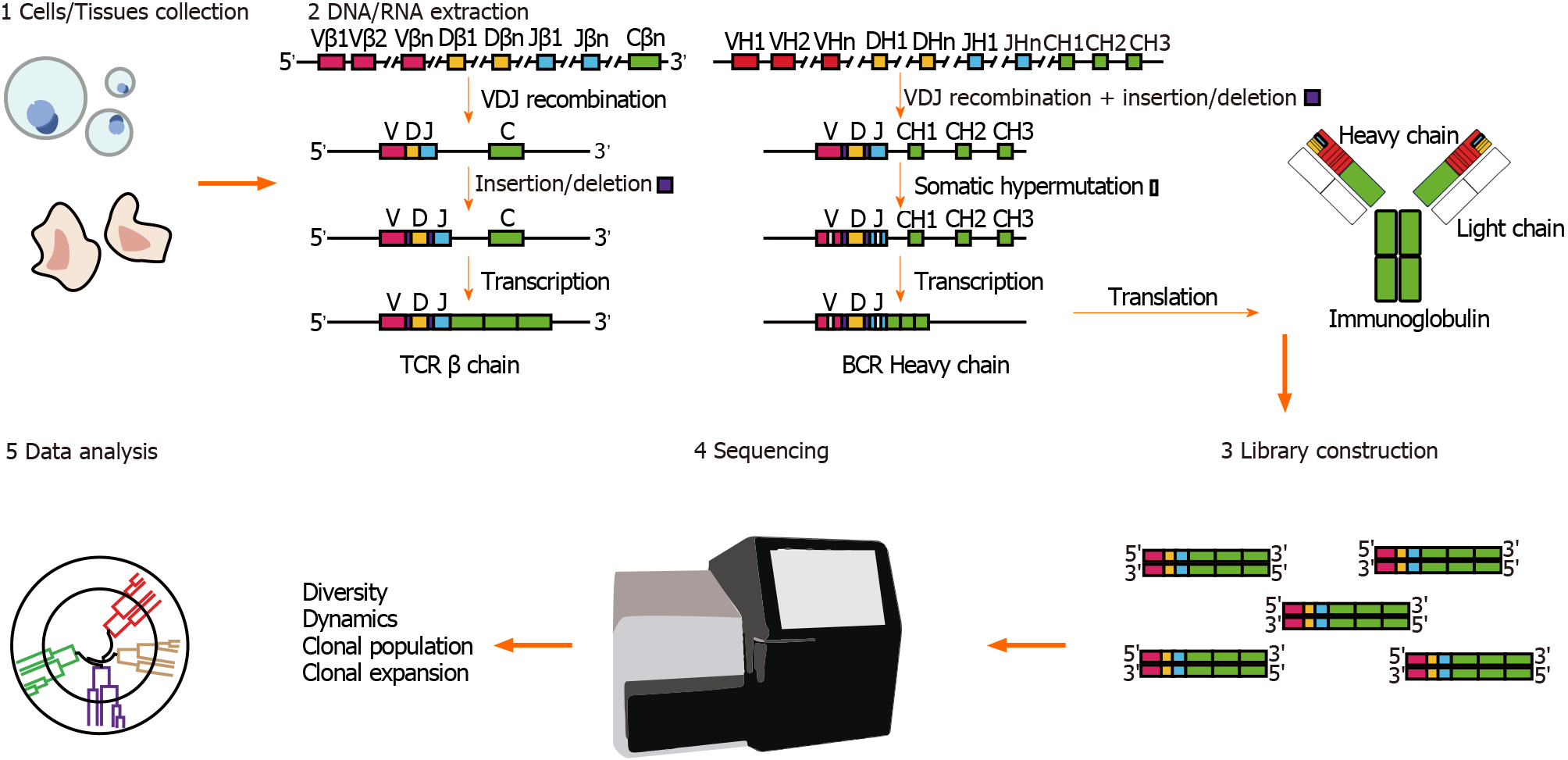
Figure 2 Workflow for immune repertoire sequencing.
There are mainly five sections in immune repertoire analysis. 1: Cell/tissue collection. 2: DNA/RNA extraction. The figure illustrates the gene recombination and transcription of T-cell receptors (TCRs) and B-cell receptors (BCRs). The V, D, and J gene segments of TCRs and BCRs undergo somatic rearrangement and gene insertion/deletion before transcription to generate highly variable CDR3 regions, which are then translated as TCRs and immunoglobulins. 3: Library construction by next-generation sequencing methods are used to complete the library preparation and amplification. 4: Data sequencing. 5: Data analysis. BCR: B-cell receptor; CDR3: Complementary determining region 3; TCR: T-cell receptor.
- Citation: Zhan Q, Xu JH, Yu YY, Lo KK E, Felicianna, El-Nezami H, Zeng Z. Human immune repertoire in hepatitis B virus infection. World J Gastroenterol 2021; 27(25): 3790-3801
- URL: https://www.wjgnet.com/1007-9327/full/v27/i25/3790.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i25.3790