Copyright
©The Author(s) 2019.
World J Gastroenterol. Nov 28, 2019; 25(44): 6508-6526
Published online Nov 28, 2019. doi: 10.3748/wjg.v25.i44.6508
Published online Nov 28, 2019. doi: 10.3748/wjg.v25.i44.6508
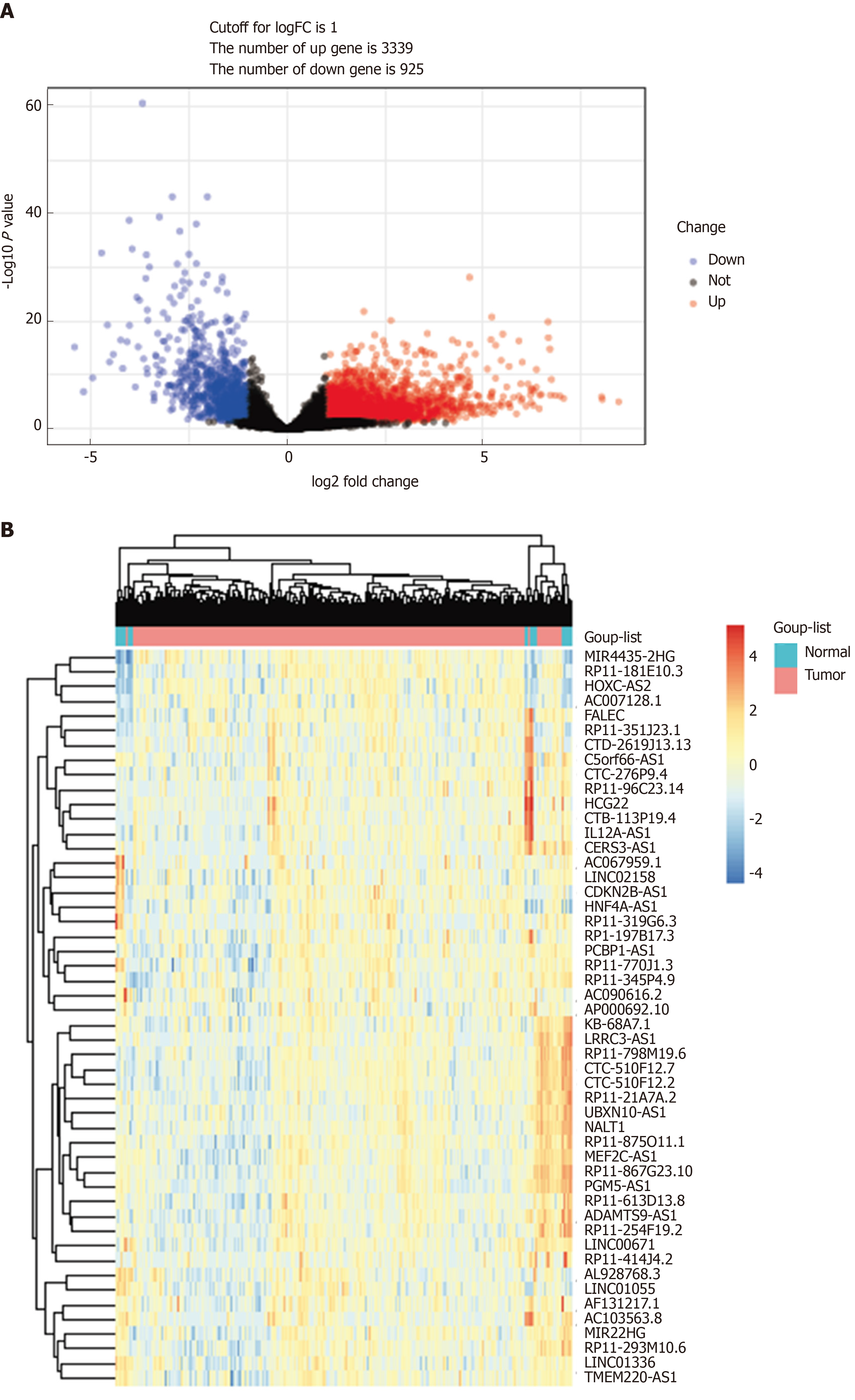
Figure 1 DELs in gastric cancer.
A: Volcano plots of the expression levels of DELs screened by limma; B: Heatmaps of the expression levels of top 50 DELs. DELs: Differentially expressed lncRNAs; lncRNAs: Long noncoding RNAs.
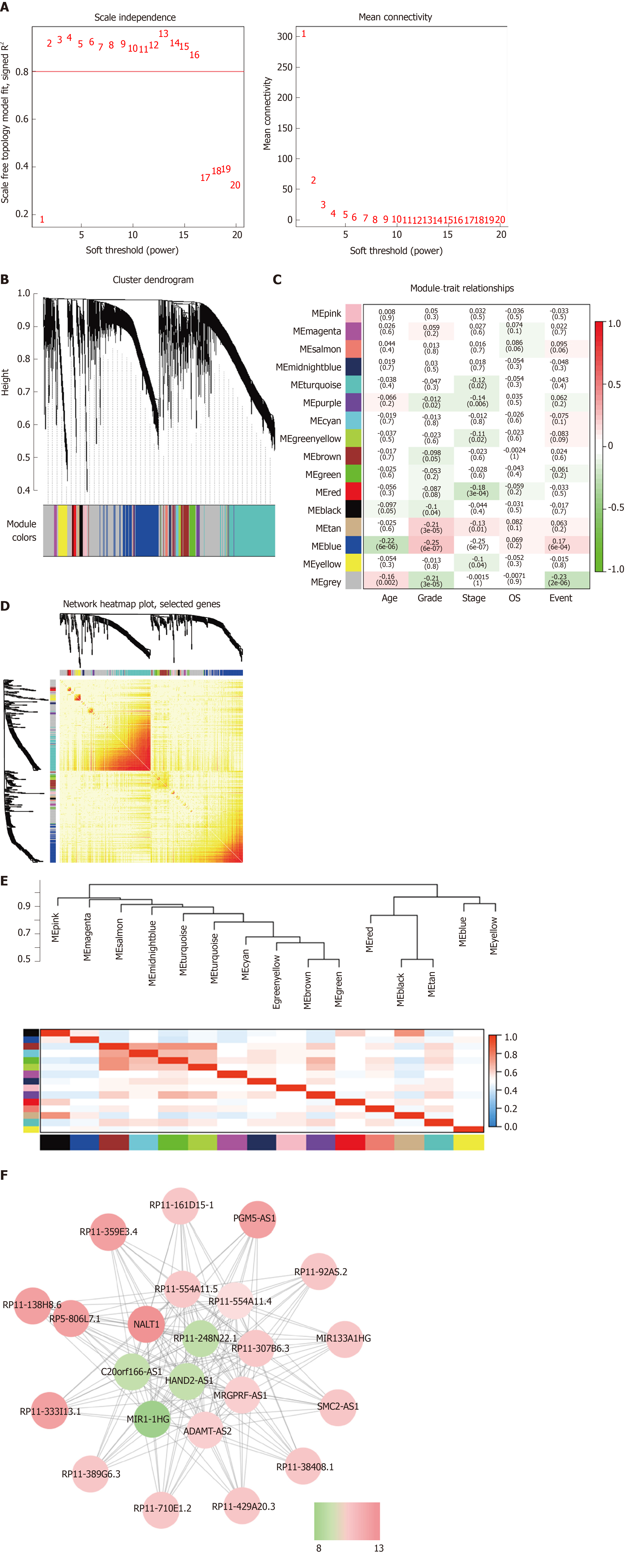
Figure 2 Weighted gene coexpression network analysis identify function module.
A: The soft-thresholding powers (β) of scale-free fit index and mean connectivity; B: Cluster dendrogram obtained by hierarchical clustering of adjacency-based dissimilarity; C: Heatmap of the correlation between module and gastric cancer clinical traits; D: Topological overlap matrix plot; E: Hierarchical clustering and heatmap of eigengene adjacency. The color bars on left and below indicate the module of each row or column; F: Weighted coexpression network for blue-module lncRNAs (Node colors are according to the molecular complex detection score). LncRNAs: Long noncoding RNAs.
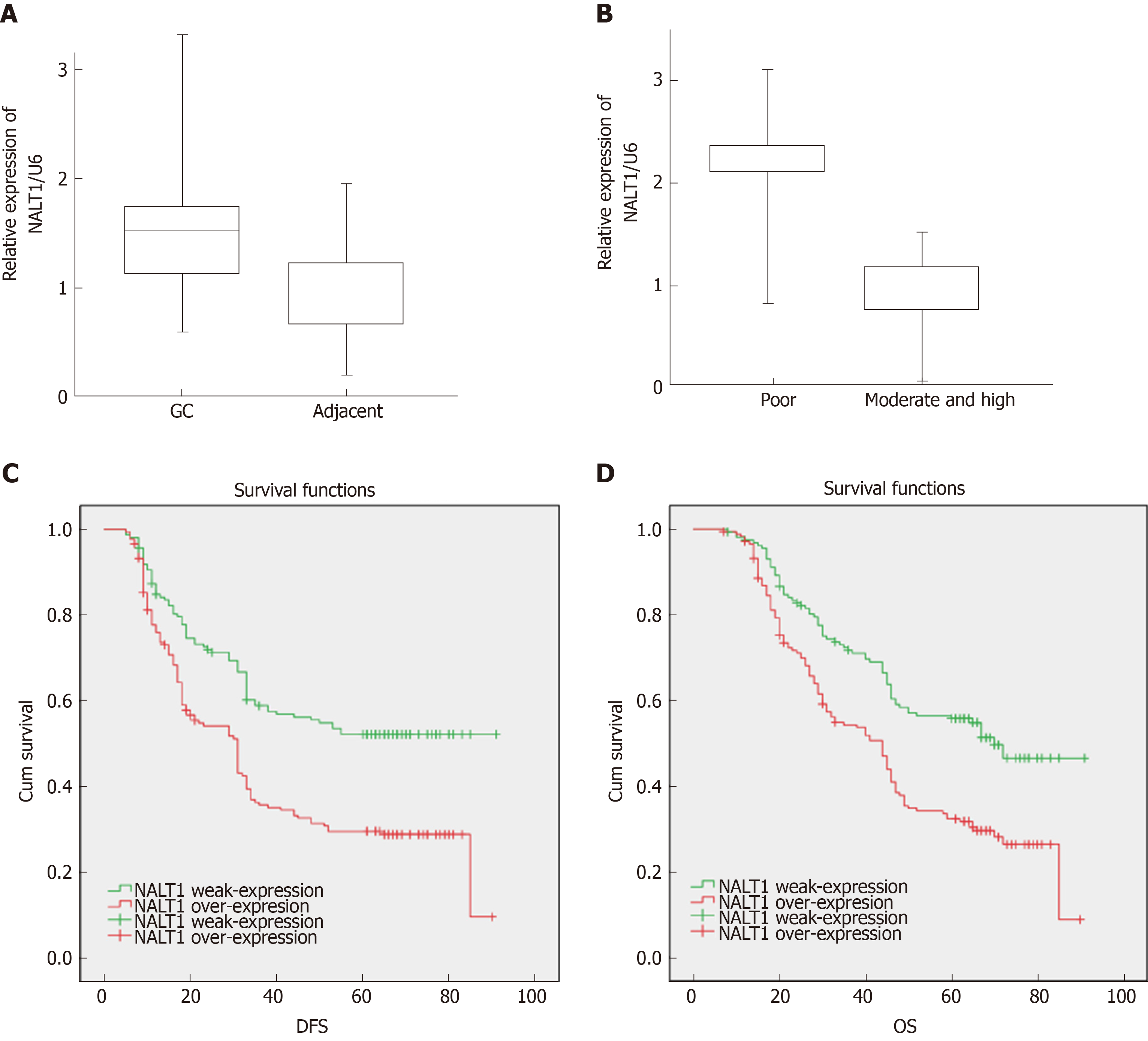
Figure 3 The expression of NALT1 in GC tissue and its effect on survival.
A: NALT1 expression in GC tissue compare to paired adjacent tissue in 35 patients; B: The relationship between NALT1 expression and tumor differentiation; C: Kaplan–Meier analysis of the correlation between NALT1 expression levels and DFS in 336 GC patients; D: Kaplan–Meier analysis of the correlation between NALT1 expression levels and OS in 336 GC patients. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer; DFS: Disease-free survival; OS: Overall survival.
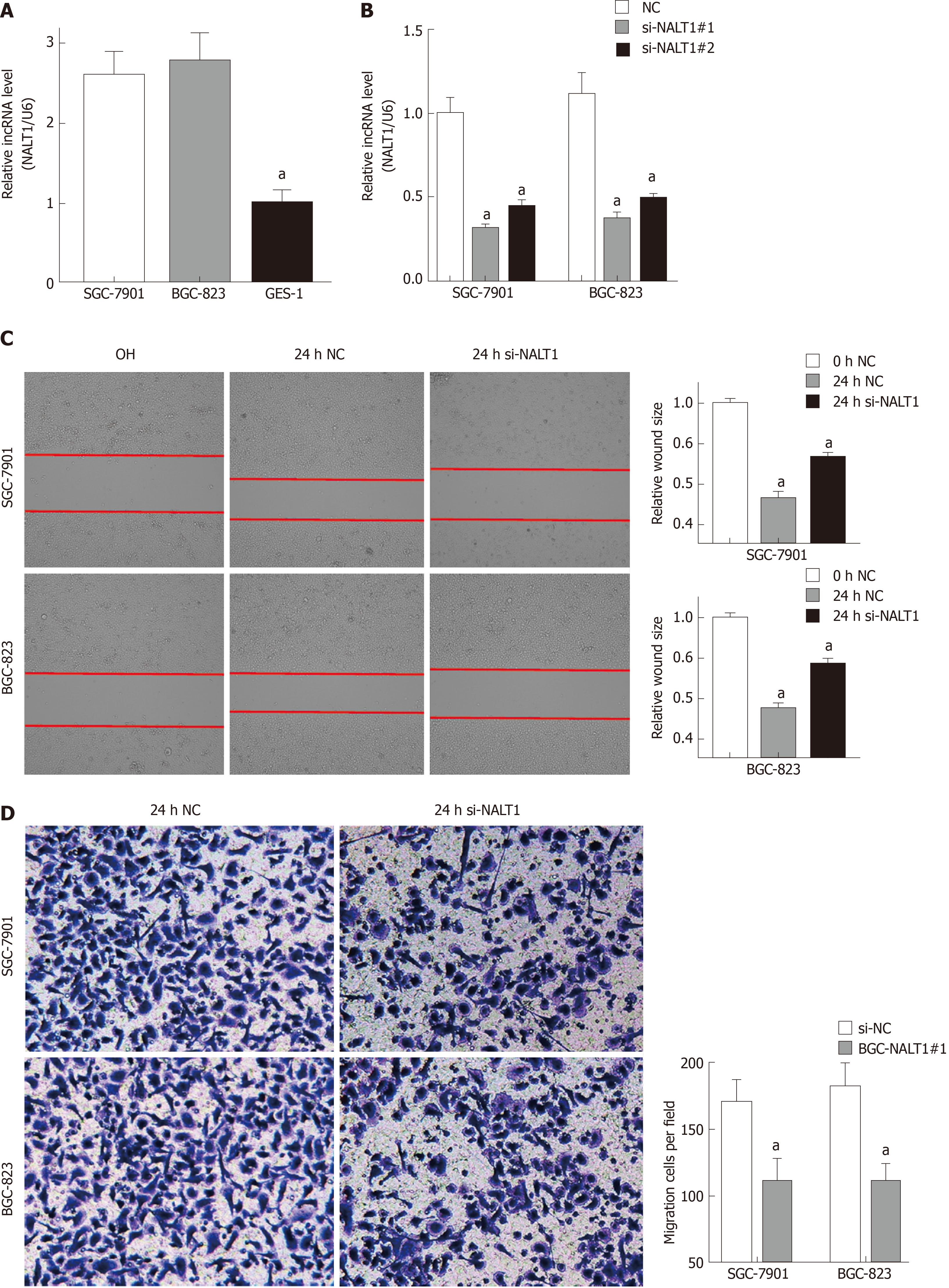
Figure 4 NALT1 induced GC invasion and metastasis.
A: NALT1 expression in GC cells compare to GES-1 cells; B: The expression of NALT1 in GC cells before and after transfection si-NALT1 sequence; C: Scrape motility assays were monitored for 24 h in NALT1-knockdown GC cells; D: Transwell assays were used to evaluate the role of NALT1 in invasion in NALT1-knockdown. U6 was used as a loading control in RT-PCR; In all figures, 100 × magnification was used, n = 3, aP < 0.05. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer; GES-1: Human normal gastric epithelial cell line.
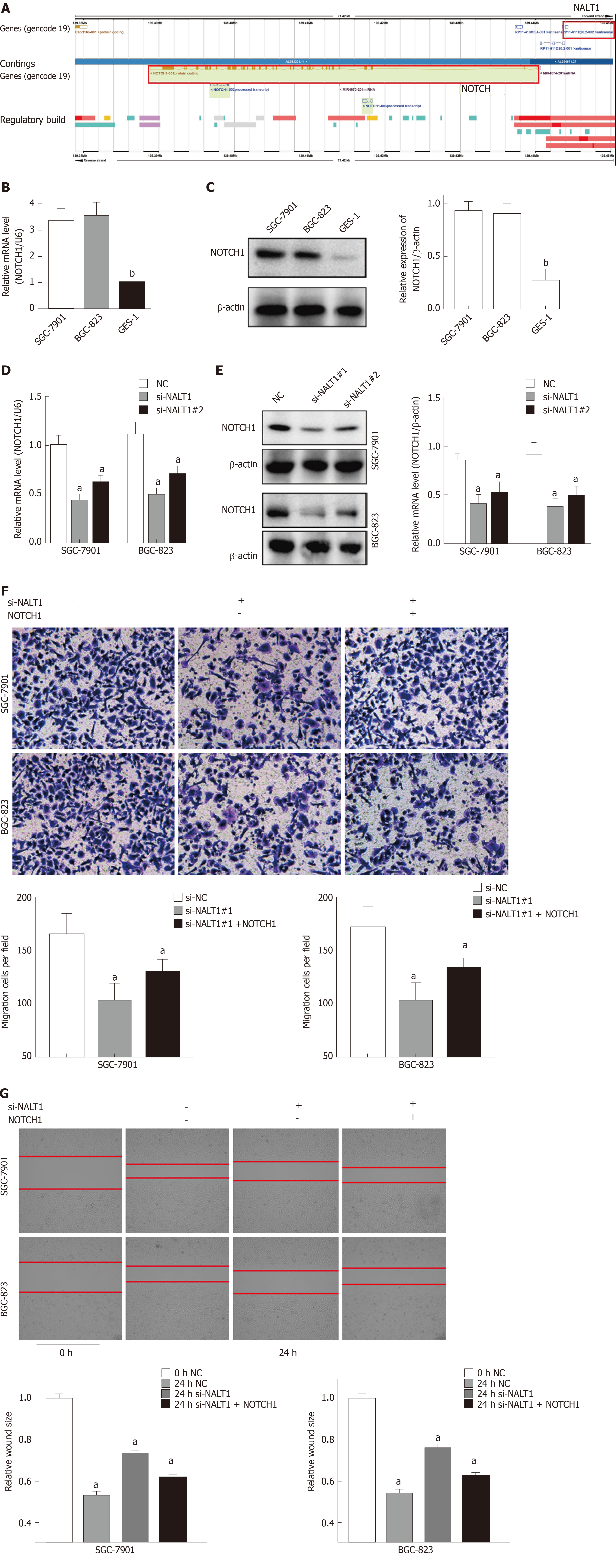
Figure 5 NALT1 influence NOTCH1 expression.
A: Schematic figure of NALT1 and NOTCH1 in chromosome 9. They were highlight with red box; B: NOTCH1 was overexpression in GC cells by RT-PCR; C: NOTCH1 was overexpression in GC cells by western-blot; D: The expression of NOTCH1 mRNA was effected by NALT1; E: The expression of protein was effected by NALT1; F: Transwell assay was used to evaluate the association of NALT1 and NOTCH1 in invasion in NALT1-knockdown and NALT1-knockdown + NOTCH1-overexpression GC cells; G: Scrape motility assays were monitored for 24 h in NALT1-knockdown and NALT1-knockdown + NOTCH1-overexpression GC cells. U6 and β-actin were used as loading control in RT-PCR and western-blot, respectively. In all figures, 100 × magnification was used. n = 3, aP < 0.05, bP < 0.01. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer.
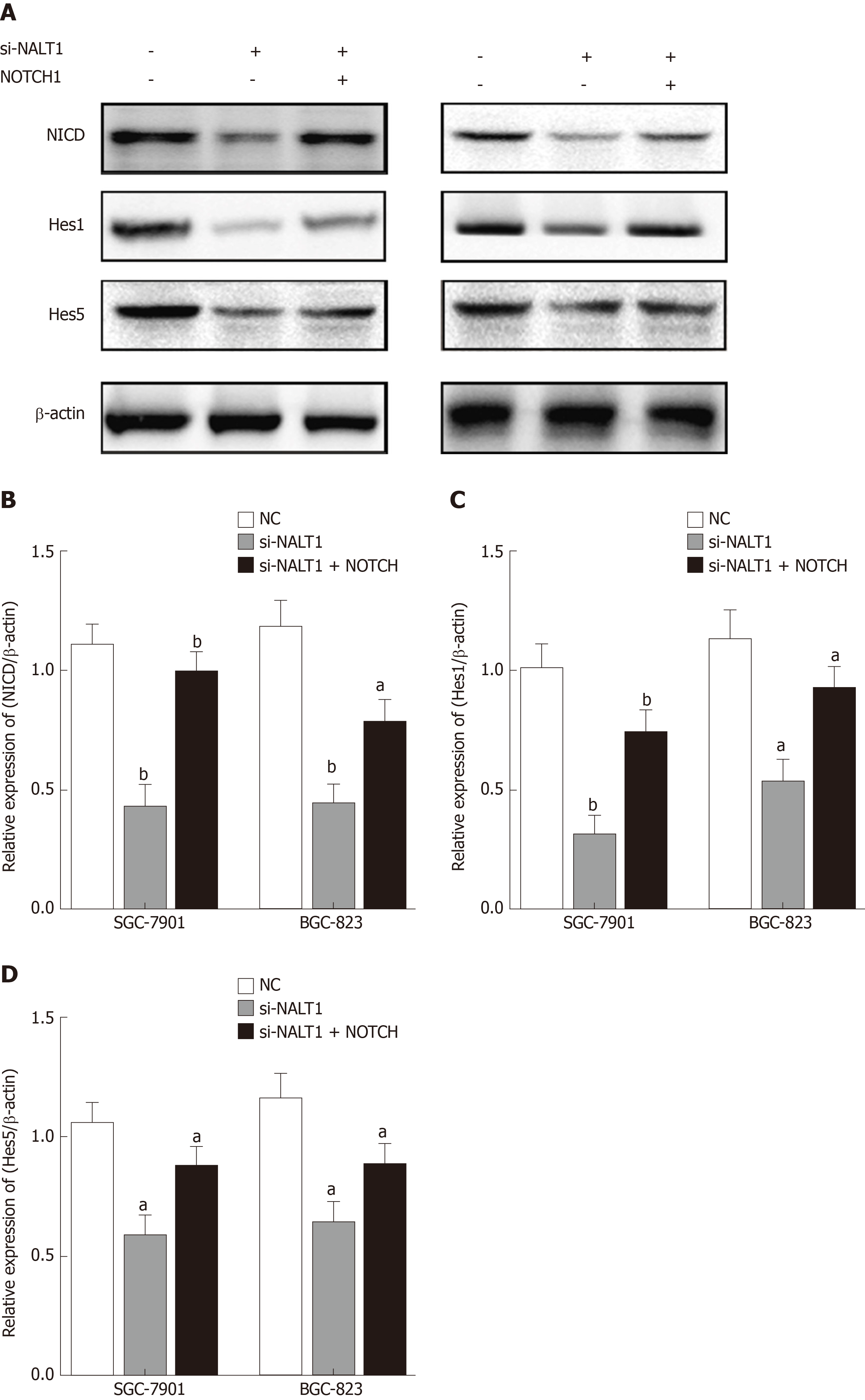
Figure 6 Decreased NALT1 suppressed the NOTCH signaling pathway.
A: Western blot was used to detect whether the loss of NALT1 led to the abnormal expression of NOTCH signaling pathway. NICD, Hes1, and Hes5 were selected; B: The relative expression of NICD; C: The relative expression of Hes1; D: The relative expression of Hes5. β-actin was used as a loading control; n = 3, aP < 0.05, bP < 0.01. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1.
- Citation: Piao HY, Guo S, Wang Y, Zhang J. Long noncoding RNA NALT1-induced gastric cancer invasion and metastasis via NOTCH signaling pathway. World J Gastroenterol 2019; 25(44): 6508-6526
- URL: https://www.wjgnet.com/1007-9327/full/v25/i44/6508.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i44.6508