Copyright
©The Author(s) 2019.
World J Gastroenterol. Oct 21, 2019; 25(39): 5961-5972
Published online Oct 21, 2019. doi: 10.3748/wjg.v25.i39.5961
Published online Oct 21, 2019. doi: 10.3748/wjg.v25.i39.5961
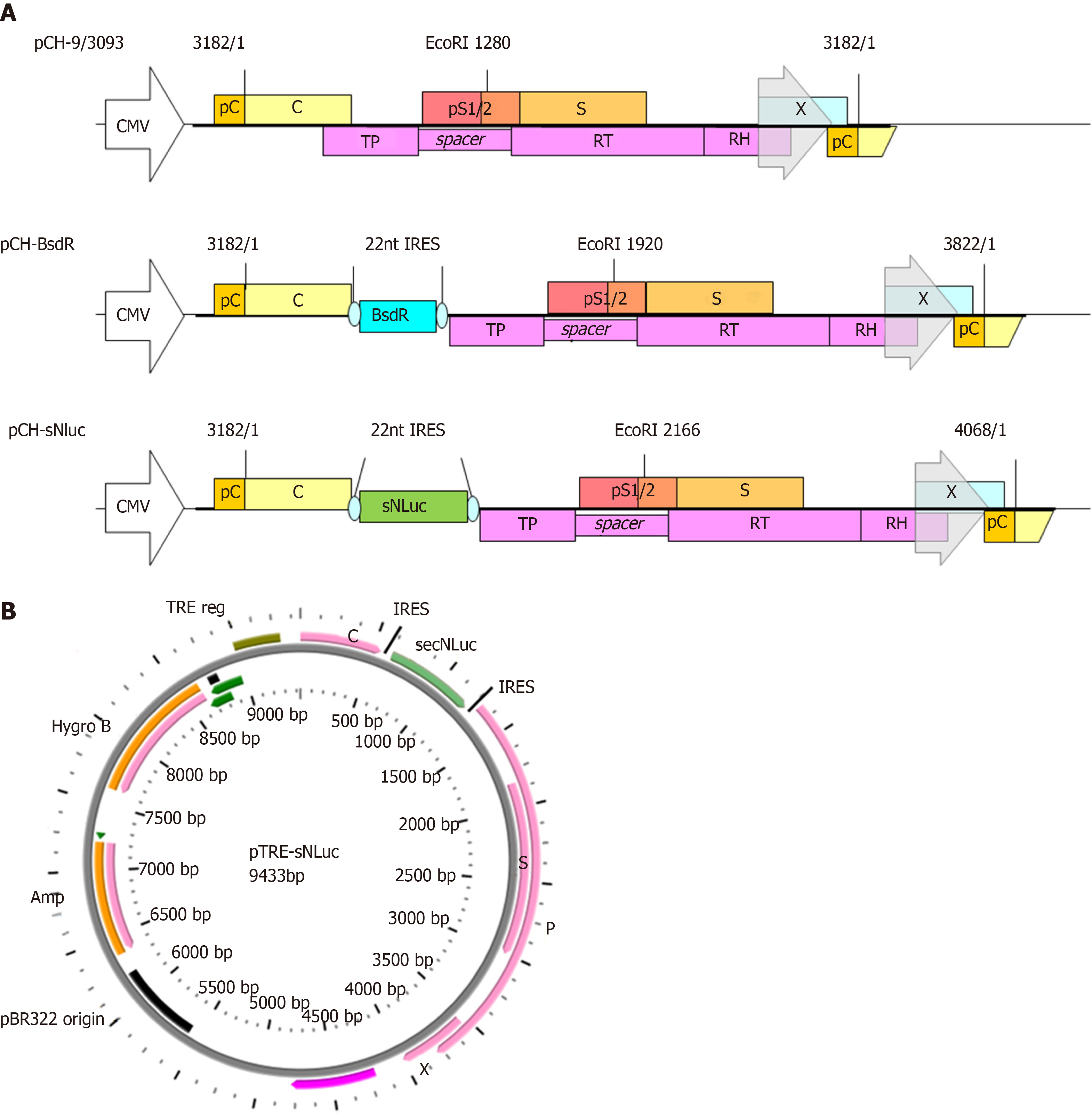
Figure 1 Genomic organization of wild-type hepatitis B virus vector pCH-3093 and replication-competent hepatitis B virus vectors.
A: The parental plasmid pCH-3093 is based on hepatitis B virus (HBV) genotype D, subtype ayw (GenBank accession No. V01460.1), including the 1.056 HBV genome, in which the CMV promoter replaces the primitive HBV core promoter to start pregenomic RNA (pgRNA) transcription. The HBV genome contains four open reading frames (ORFs): PreC/C (encoding precore protein giving rise to the hepatitis B e antigen and core protein), pS1/2 and S (encoding preS1, preS2, and S domains of the envelope proteins, respectively), X (encoding hepatitis B x antigen), and P (encoding viral polymerase, Pol). All regions are widely overlapping, and the P ORF overlaps with all other ORFs. TP, RT, and RH indicate terminal protein, reverse transcriptase, and RNase H domains of Pol (P). Regarding pCH-BsdR, the BsdR gene is inserted among the uncoupled P ORFs from the preC/C ORFs of pCH-3093. Regarding pCH-secNLuc, secNLuc gene was used to replace BsdR gene and inserted into among uncoupled P ORFs from the preC/C ORF of pCH-BsdR; B: Schematic map of pTRE-sNLuc vector. pTRE-sNLuc is based on pTRE-HBV-C7-5 and pTRE-HBVT, in which the Tet responsive promoter replaces the primitive HBV core promoter to control HBV pgRNA transcription. A hygromycin resistance gene serves as a selection marker, and the HBV-sNLuc fragment stems from pCH-secNLuc. HBV: Hepatitis B virus; TRE: Tet responsive promoter; ORFs: Open reading frames; pgRNA: Pregenomic RNA; PreC/C: Encoding precore protein giving rise to the hepatitis B e antigen and core protein; S: Encoding preS1, preS2, and S domains of the envelope proteins, respectively; X: Encoding hepatitis B x antigen; P: Encoding viral polymerase, Pol.
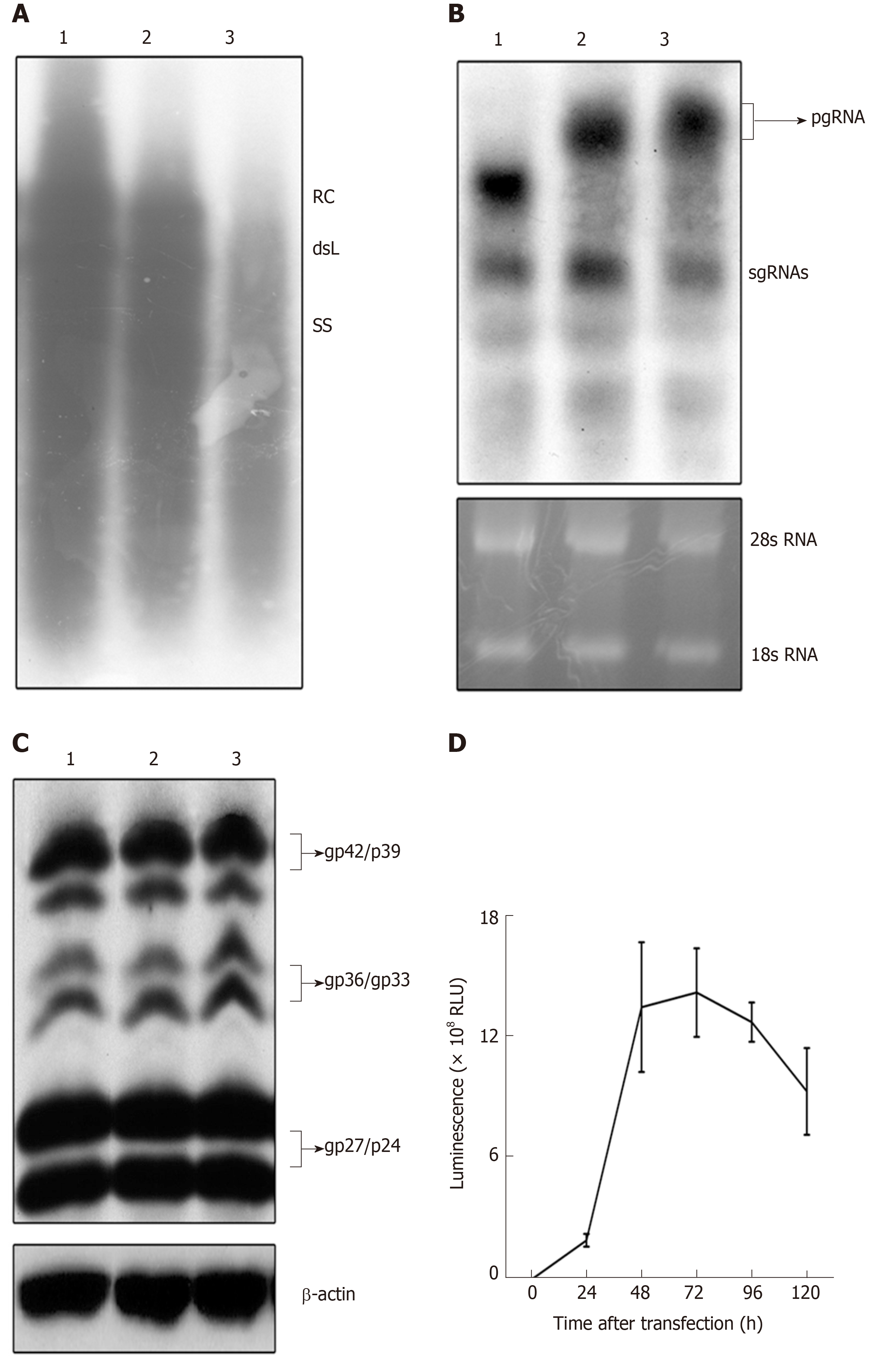
Figure 2 Detection of expression and replication of the recombinant hepatitis B virus vectors.
HepG2 cells were transfected with pCH-3093 (1), pCH-BsdR (2), and pCH-sNLuc (3). A: Replication efficiency of HBV vectors. Cytoplasmic HBV DNA was detected at 4 d post-transfection, and replicative intermediates were monitored by Southern blot using a 32P-labeled HBV probe. The positions of relaxed circular and double-stranded linear DNA are indicated; B: Detection of the RNA transcription of HBV vectors. Total intracellular RNA was analyzed by Northern blot with a 32P-labeled HBV-specific probe. The positions of the pregenomic RNA and subgenomic RNAs are indicated, and 28S and 18S rRNA bands are shown as loading controls; C: Detection of envelope protein expression of HBV vectors. Western blot analysis was performed using a 4/7B HBsAg antibody. The bands correspond to the L protein (gp42/p39), M protein (gp36/gp33), and S protein (gp27/p24), and β-actin was used as a loading control; D: The dynamic luciferase activity of pCH-sNLuc. The vector pCH-sNLuc was transiently transfected into HepG2 cells, and luciferase expression in the supernatant was detected at the indicated time points (24, 48, 72, 96, and 120 h). HBV: Hepatitis B virus; RLU: Relative light unit; dsL: Double-stranded linear; RC: Relaxed circular; pgRNA: Pregenomic RNA; sgRNAs: Subgenomic RNAs.
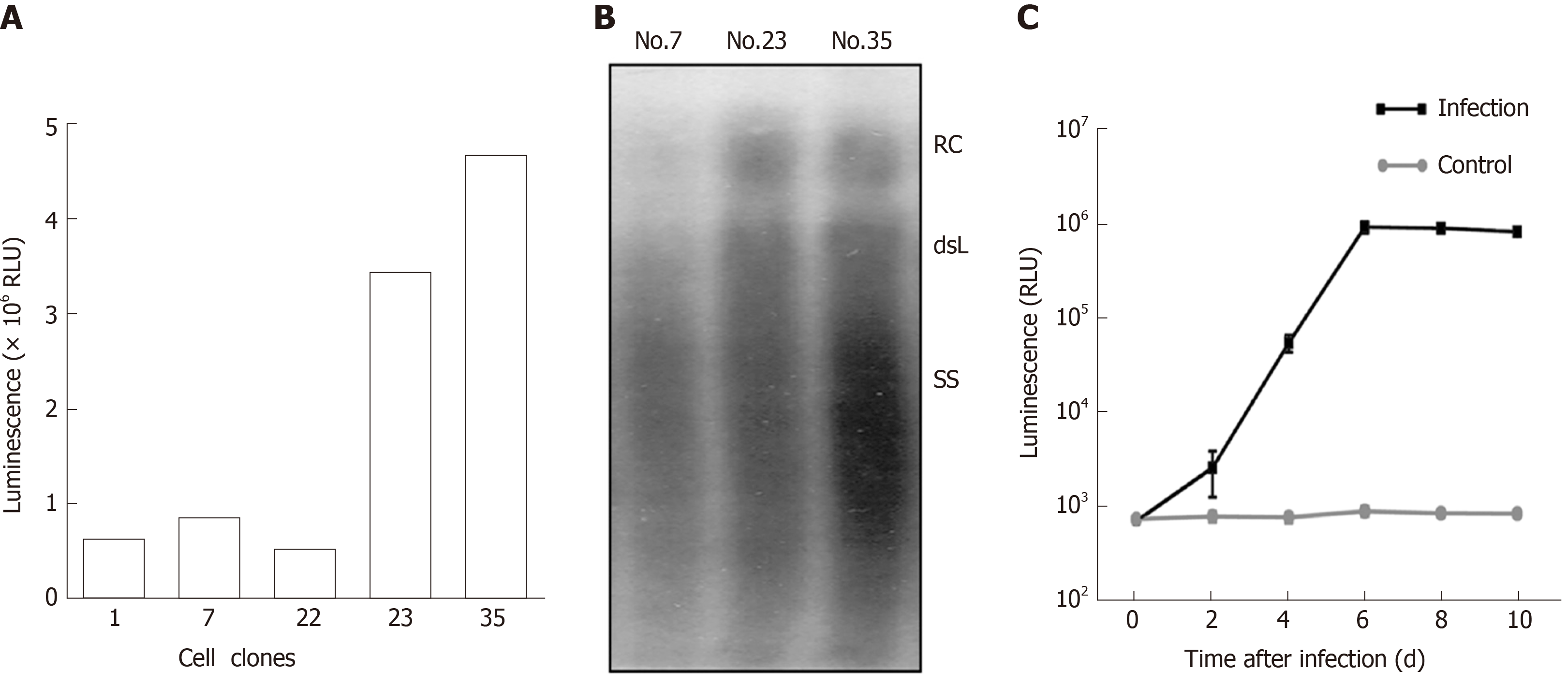
Figure 3 Establishment of secretory Nanoluc Luciferase reporter gene cell lines carrying stably secreted hepatitis B virus particles.
HepG2 TA-7 cells were transfected with the replication-competent vector pTRE-sNLuc, and the isolated clone of cells was selected in the presence of hygromycin. A: Selection of cell lines with good luciferase activity. After several passages, the Nos. 1, 7, 22, 23, and 35 cell lines were screened, and luciferase expression in the supernatant was detected with Nano-Glo luciferase assay reagent; B: Selection of cell lines with good hepatitis B virus (HBV) replication. Extracellular viral particles of cell lines Nos. 7, 23, and 35 were analyzed by Southern blot using a 32P-labeled HBV probe. The relaxed circular DNA and double-stranded linear DNA signals are indicated; C: Infection of HepaRG cells by secretory Nanoluc Luciferase recombinant HBV particles. Differentiated HepaRG cells were inoculated with viral particles from supernatants of cell lines No. 35 or not (negative control), and luciferase expression in the supernatant was detected at the indicated time points (0, 2, 4, 6, 8, and 10 d). HBV: Hepatitis B virus; RLU: Relative light unit.
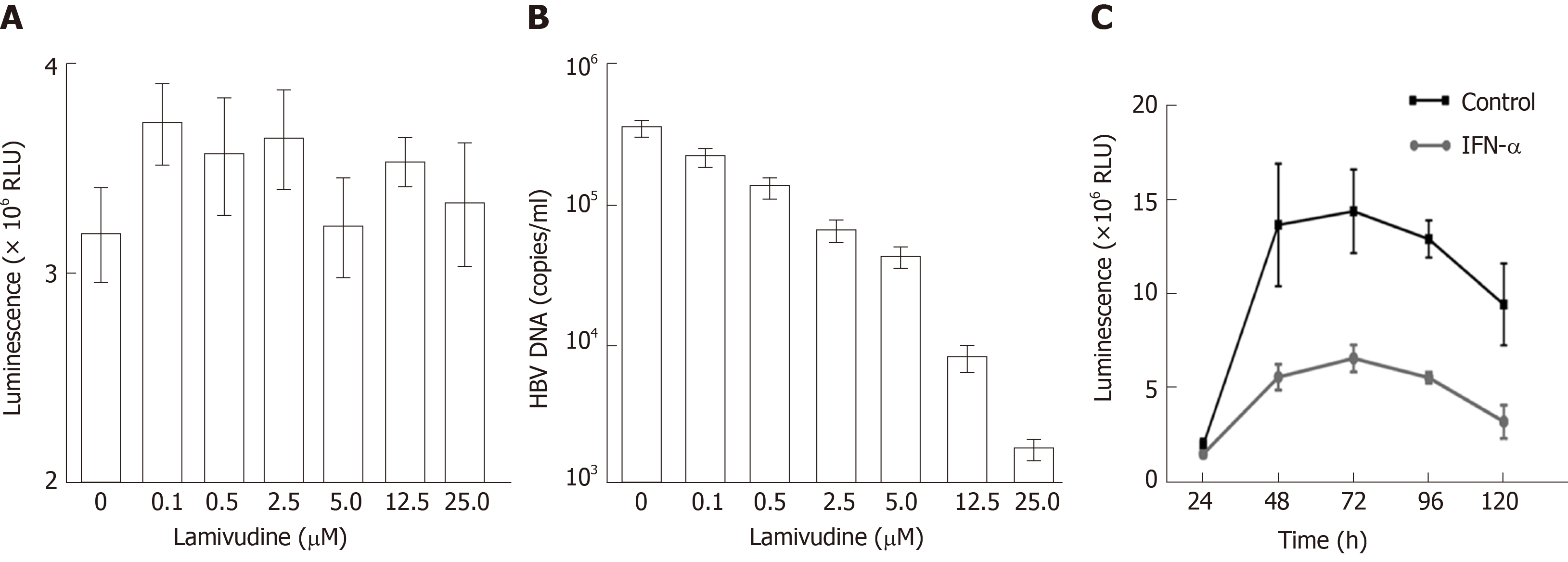
Figure 4 Drug susceptibility testing for the new hepatitis B virus -NLuc-35 cell lines.
A: Effect of lamivudine on luciferase expression in hepatitis B virus (HBV)-NLuc-35 cell lines. HBV-NLuc-35 cell lines were treated with increasing concentrations of lamivudine (0, 0.1, 0.5, 2.5, 5.0, 12.5, and 25.0 µM). After 72 h, luciferase expression in the supernatant was detected with Nano-Glo luciferase assay reagent; B: Cell line-borne HBV affected by lamivudine. The number of copies of HBV DNA in the supernatant was measured by qPCR; C: Effect of IFN-α on HBV-NLuc-35 cell lines. HBV-NLuc-35 cell lines were inoculated with 3000 IU/mL IFN-α (down line) or untreated (up line), and luciferase expression in the supernatant was detected at the indicated time points (24, 48, 72, 96, and 120 h). HBV: Hepatitis B virus; RUL: Relative light unit.
- Citation: Ruan J, Ping CY, Sun S, Cheng X, Han PY, Zhang YG, Sun DX. Construction of a replication-competent hepatitis B virus vector carrying secreted luciferase transgene and establishment of new hepatitis B virus replication and expression cell lines. World J Gastroenterol 2019; 25(39): 5961-5972
- URL: https://www.wjgnet.com/1007-9327/full/v25/i39/5961.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i39.5961