Copyright
©The Author(s) 2019.
World J Gastroenterol. Aug 21, 2019; 25(31): 4300-4319
Published online Aug 21, 2019. doi: 10.3748/wjg.v25.i31.4300
Published online Aug 21, 2019. doi: 10.3748/wjg.v25.i31.4300
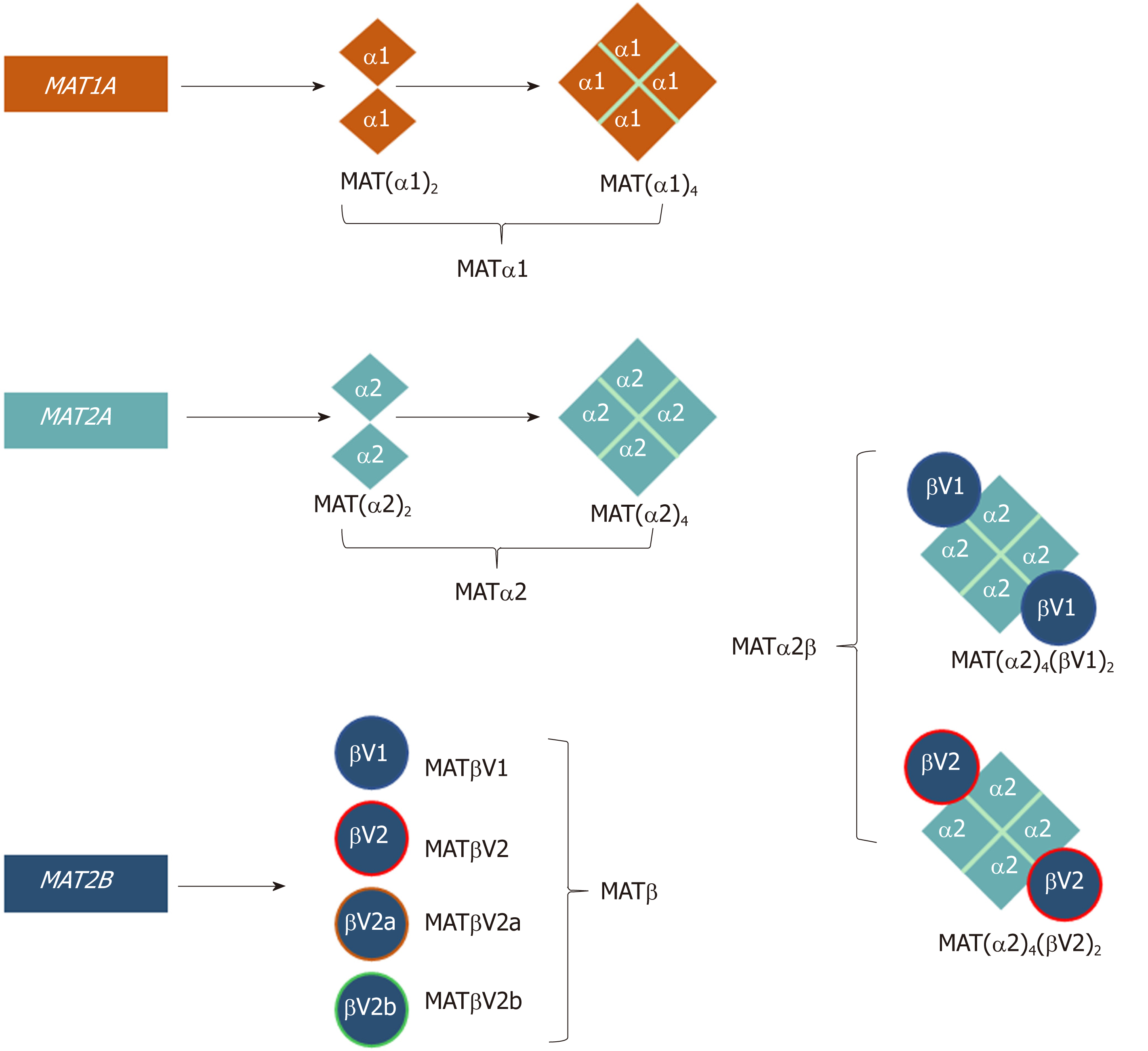
Figure 1 Schematic representation of the oligomeric states of mammalian MAT enzymes.
MAT1A and MAT2A genes encode the catalytic subunits MATα1 and MATα2, respectively. Both MATα1 and MATα2 can be organized as dimers and tetramers. MAT2B encodes the regulatory subunit for which there are four isoforms, MATβV1, MATβV2, MATβV2a, and MATβV2b with the former two being the major splice variants. MATα2 and MATβ interact to give rise to the MATα2β complexes. The MAT(α2)4(βV1)2 and MAT(α2)4(βV2)2 complexes consist of a MATα2 tetramer flanked by two MATβV1 or MATβV2 subunits.
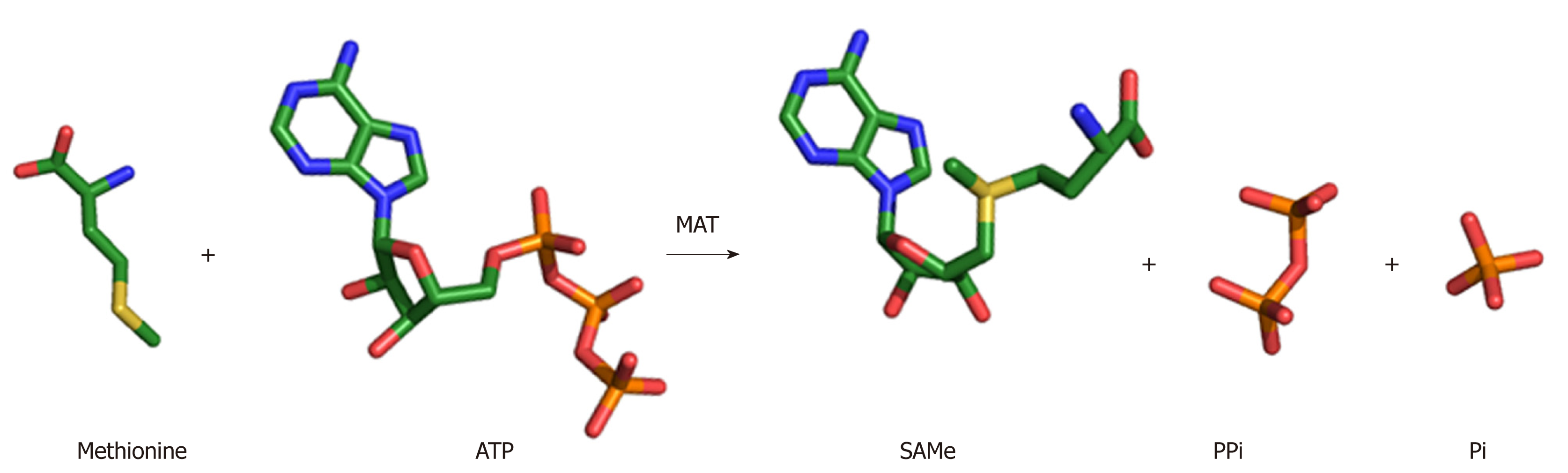
Figure 2 SAMe synthesis reaction.
MAT enzyme catalyzes the biosynthesis of SAMe from the amino acid methionine and the energy molecule ATP. The sulphur atom of methionine attacks the C5’ atom of ATP displacing the tripolyphosphate (PPPi) moiety to form SAMe. The PPPi is then hydrolyzed giving rise to pyrophosphate (PPi) and orthophosphate (Pi).
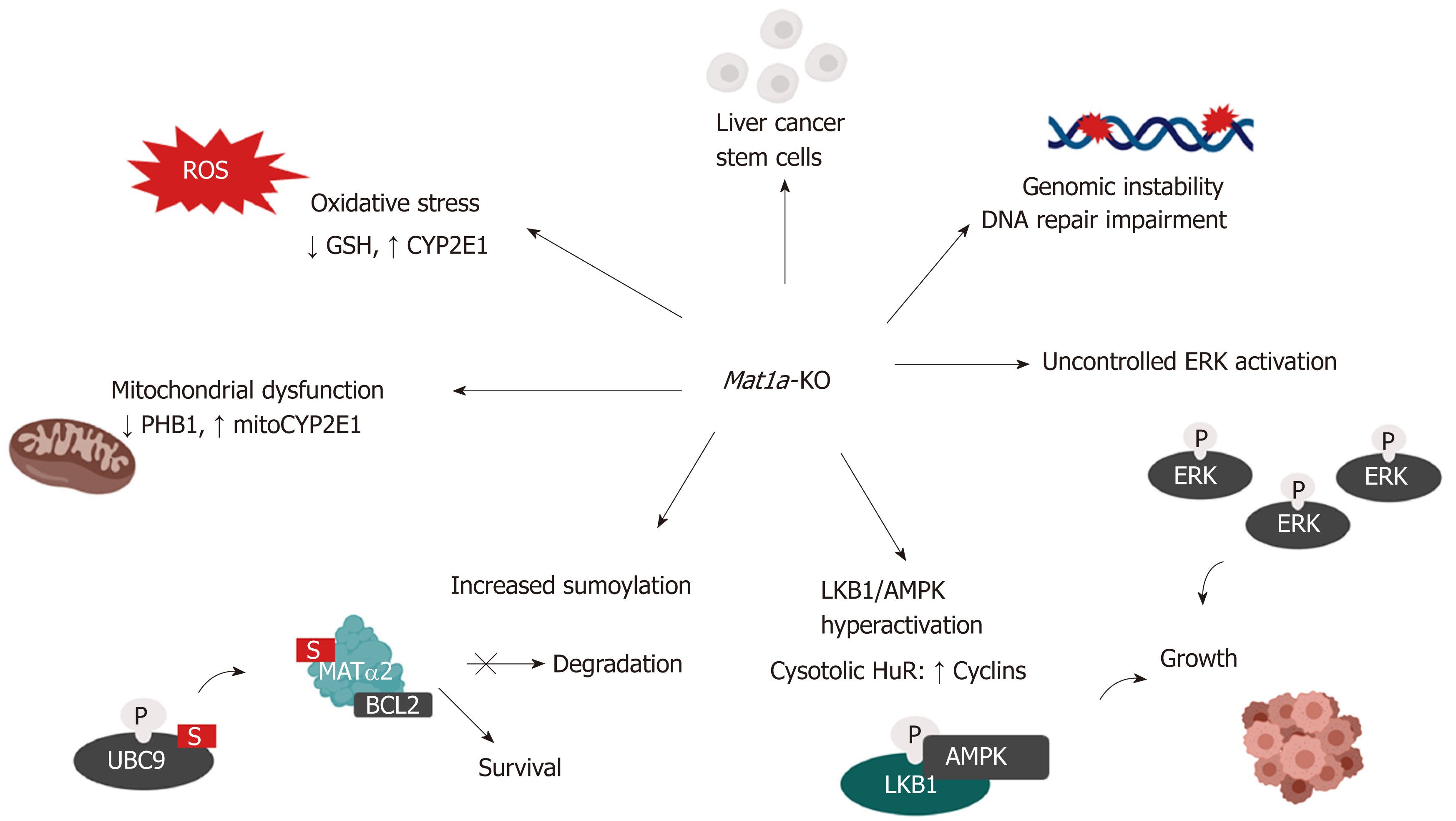
Figure 3 Mechanisms of hepatocellular carcinoma development in the Mat1a knockout mouse.
Multiple mechanisms are known to influence hepatocellular carcinoma development in the Mat1a-KO mouse. These include: oxidative stress due to lower GSH levels and higher CYP2E1 expression; mitochondrial dysfunction due to reduced PHB1 and increased mitochondrial CYP2E1 levels; increased sumoylation, stabilizing MATα2, which acts as a transcription factor to enhance BCL-2 transcription as well as directly interacting with BCL-2 leading to its stabilization; enhanced activation of the LKB1/AMPK pathway which leads to the cytoplasmic translocation of HuR from nucleus and subsequent stabilization of cyclins; aberrant activation of ERK which promotes uncontrolled cell growth; enhanced genomic instability due to DNA hypomethylation and impaired DNA repair machinery and increased number of liver cancer stem cells with tumorigenic potential.
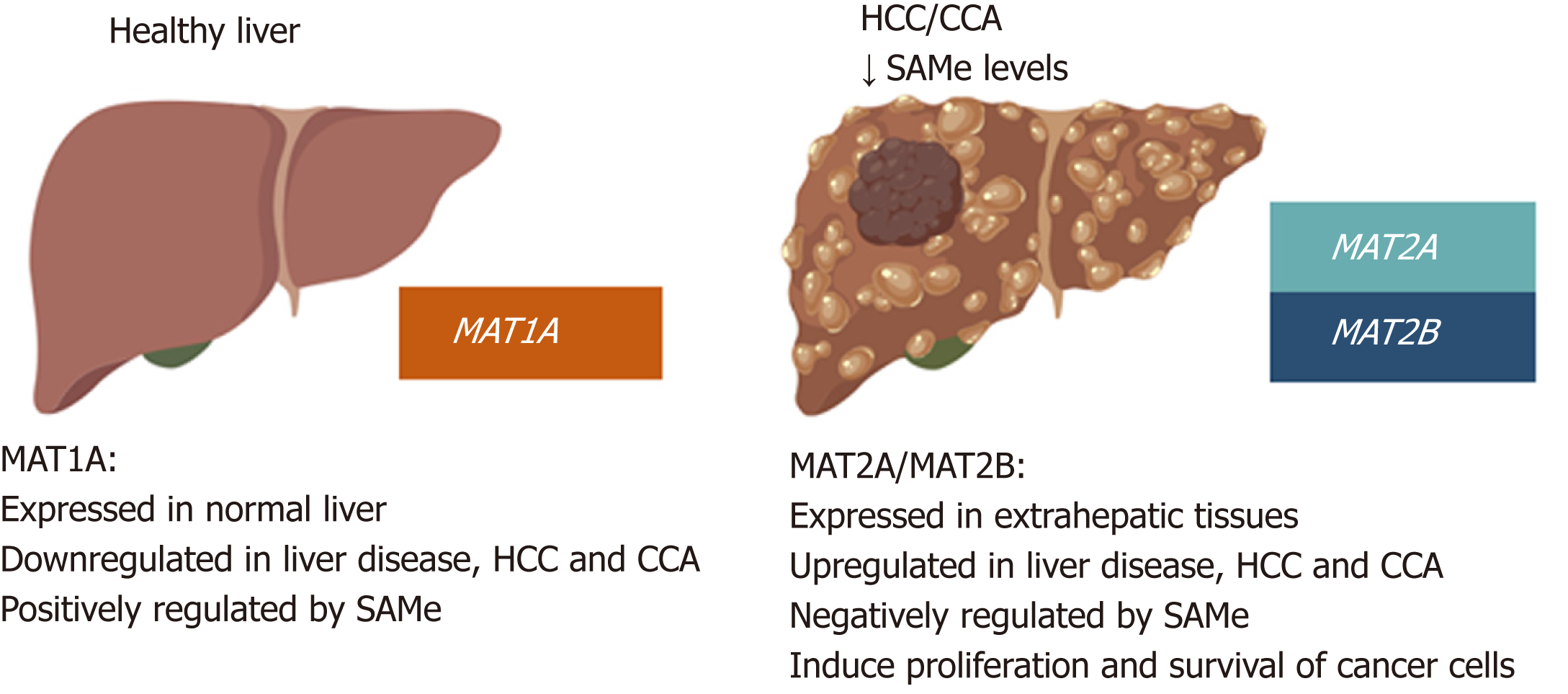
Figure 4 MAT genes expression pattern in normal liver and liver cancer.
MAT1A is mainly expressed in normal liver by hepatocytes and bile duct epithelial cells; whereas MAT2A and MAT2B are expressed in extrahepatic tissues and by the non-parenchymal cells in the liver. During liver disease and malignant transformation there is a switch from MAT1A to MAT2A/MAT2B, being MAT2A and MAT2B the predominant MAT genes expressed in HCC and CCA. MAT1A maintains the differentiated state of the liver whilst MAT2A and MAT2B give proliferative and survival advantages to cancer cells. SAMe positively regulates MAT1A and negatively regulates MAT2A and MAT2B as well as feedback inhibits MATII. Accordingly, steady state SAMe levels are lower in patients with chronic liver disease and liver cancer due to the switch from MAT1A to MAT2A/MAT2B.
- Citation: Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J Gastroenterol 2019; 25(31): 4300-4319
- URL: https://www.wjgnet.com/1007-9327/full/v25/i31/4300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i31.4300