Copyright
©The Author(s) 2019.
World J Gastroenterol. May 7, 2019; 25(17): 2086-2098
Published online May 7, 2019. doi: 10.3748/wjg.v25.i17.2086
Published online May 7, 2019. doi: 10.3748/wjg.v25.i17.2086
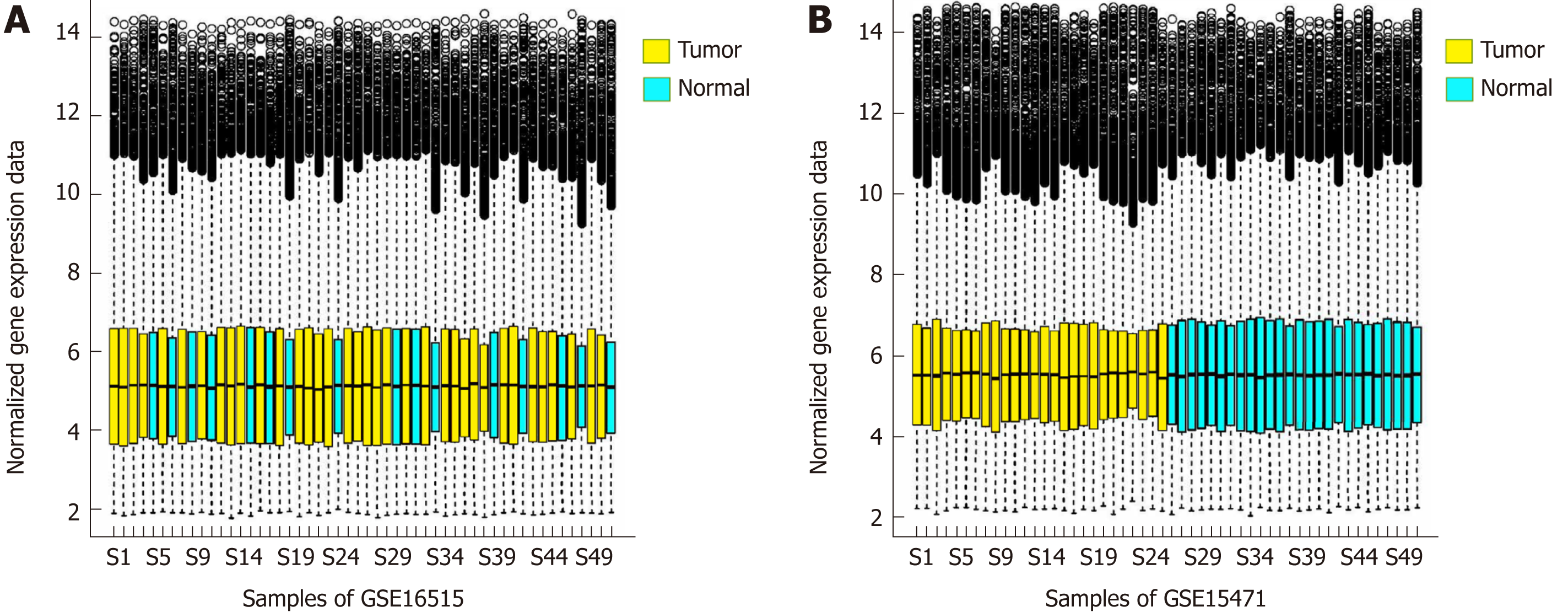
Figure 1 Box diagram of the gene expression distribution.
A: Box diagram of the gene expression distribution of each sample after the standardization of set GSE16515. B: Box diagram of the gene expression distribution of each sample after the standardization of set GSE15471.
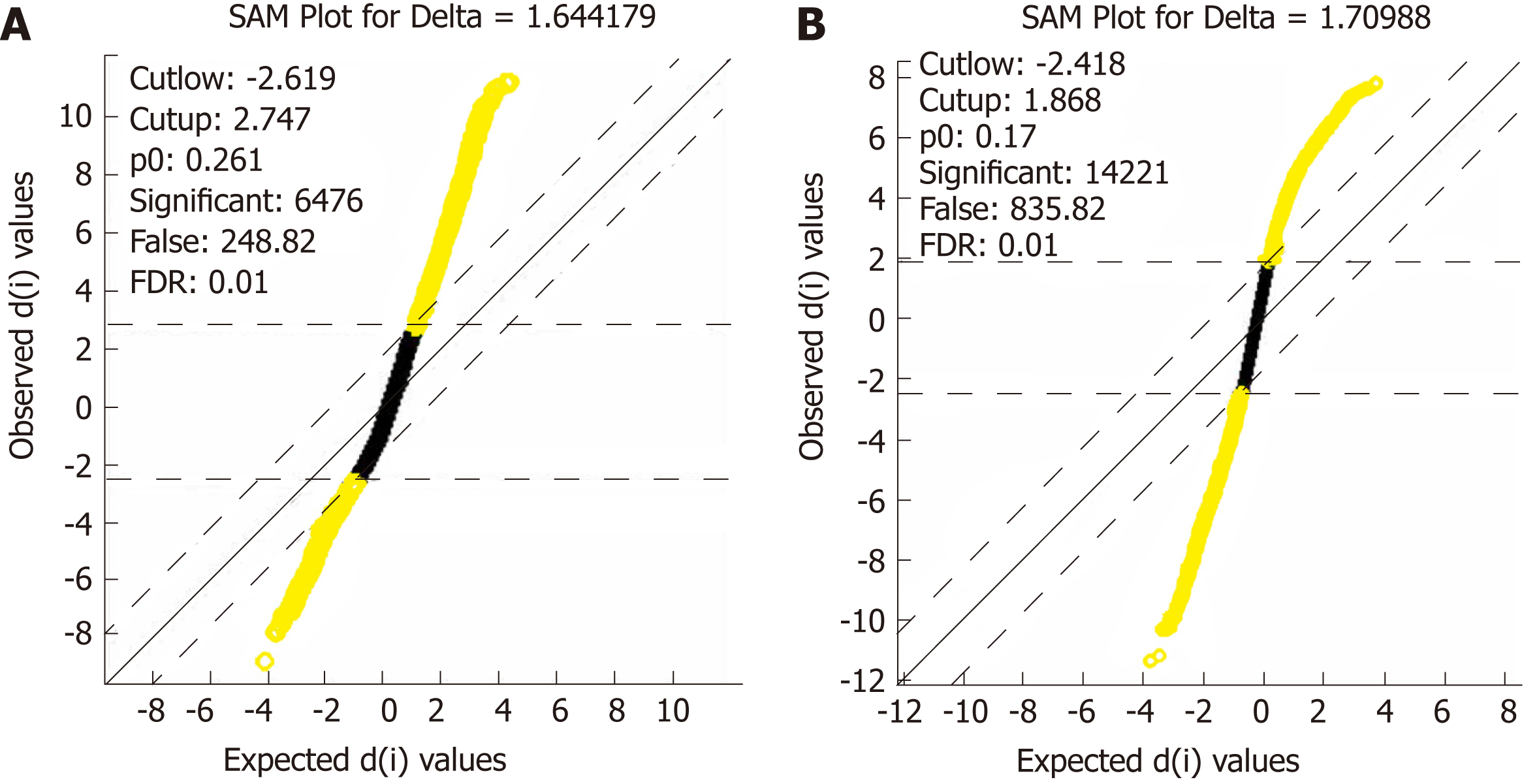
Figure 2 Distribution diagram of the statistical analysis of gene expression.
A: Distribution diagram of the statistical analysis of gene expression after the extraction of differentially expressed genes in set GSE16515; B: Distribution diagram of the statistical analysis of gene expression after the extraction of differentially expressed genes in set GSE15471. Yellow for differentially expressed genes and black for non-differentially expressed genes.
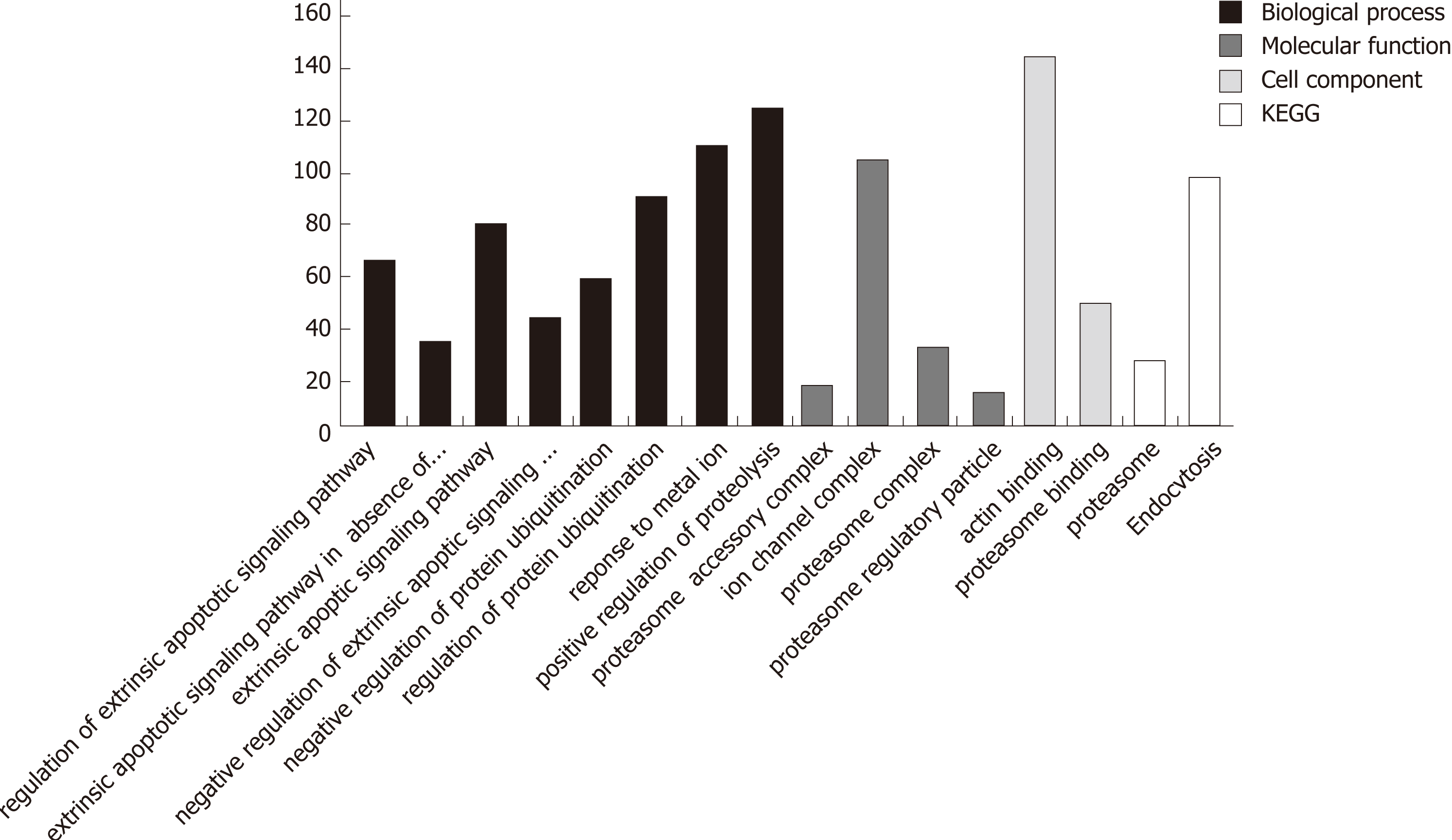
Figure 3 Distribution of the numbers of differentially expressed genes in functional items associated with apoptosis/autophagy.
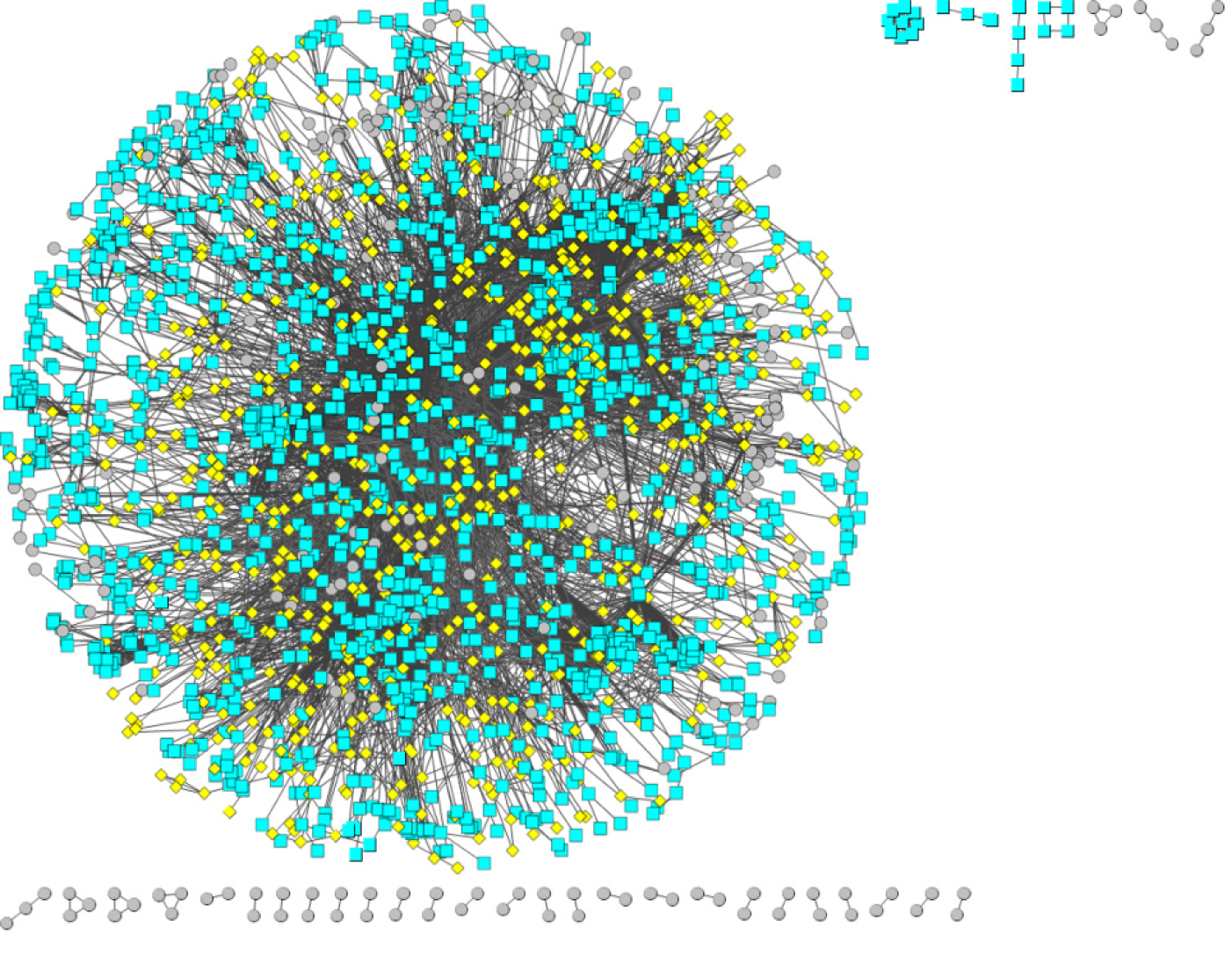
Figure 4 Protein interaction network composed of differentially expressed genes.
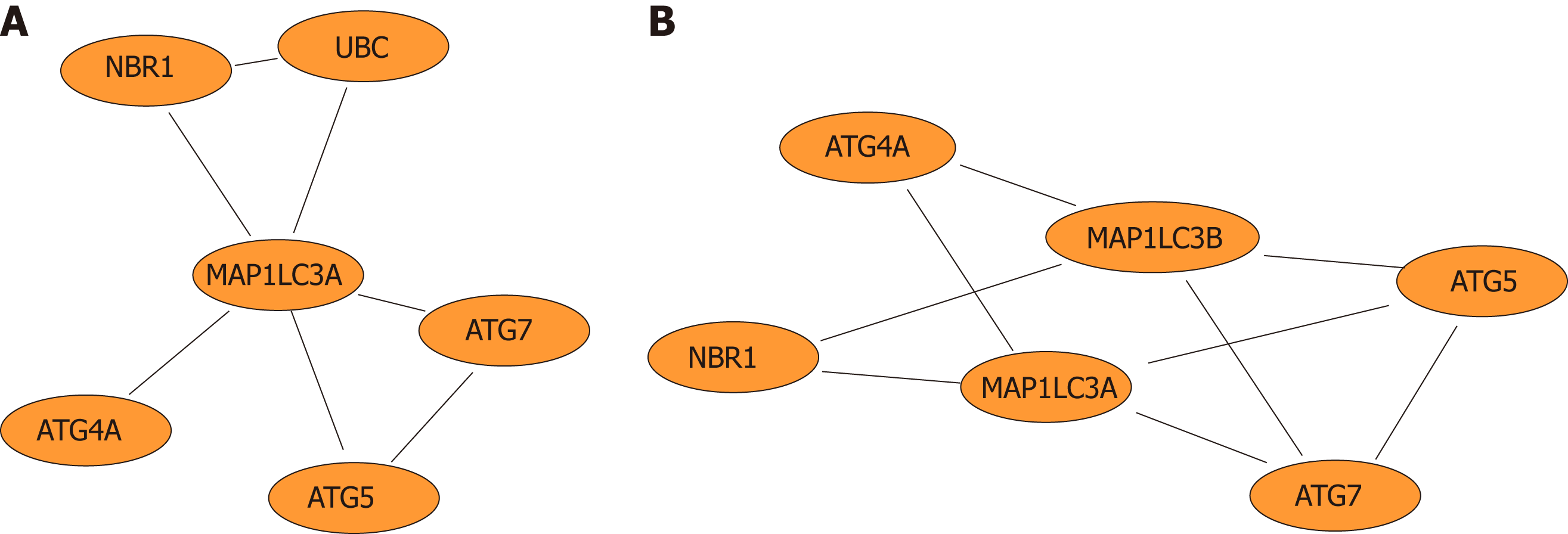
Figure 5 Network diagram.
A: Network diagram of genes that directly interact with the autophagy-related protein microtubule-associated protein 1A/1B-light chain 3 (LC3) gene; B: Network diagram of functional modules in which the LC3 gene is involved.
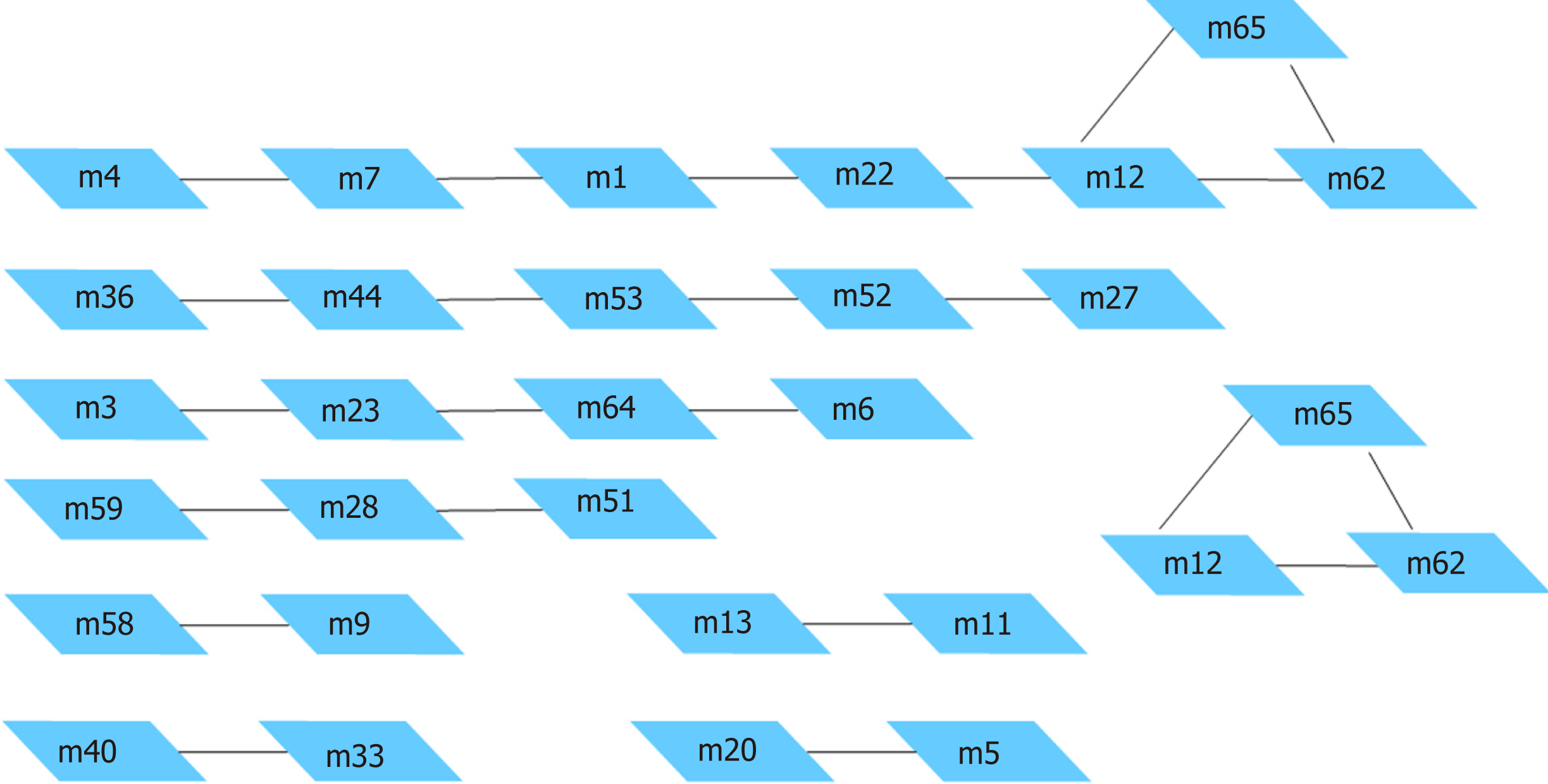
Figure 6 Module pairs with significant crosstalk.
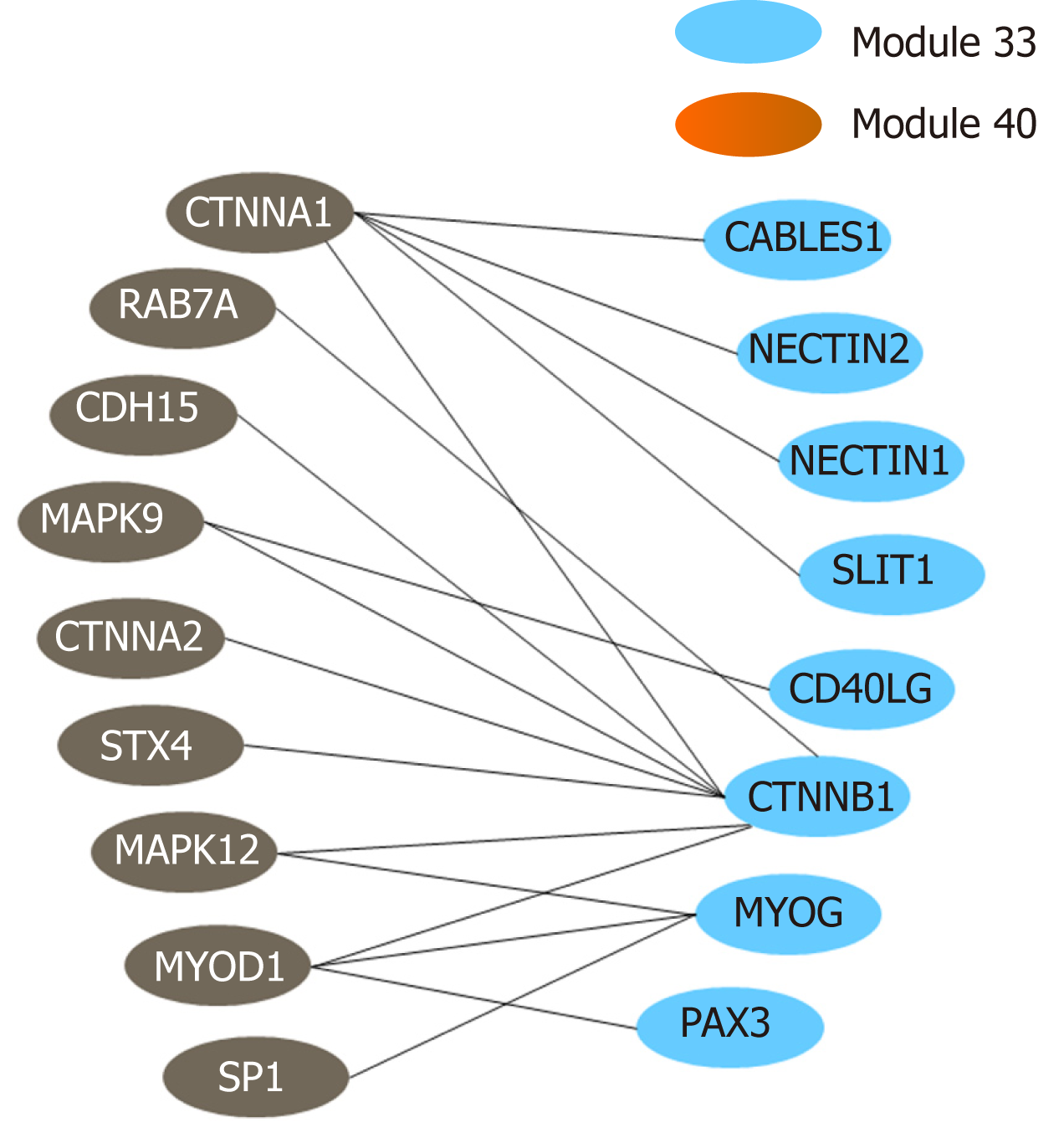
Figure 7 Crosstalk relationship between modules 33 and 44.
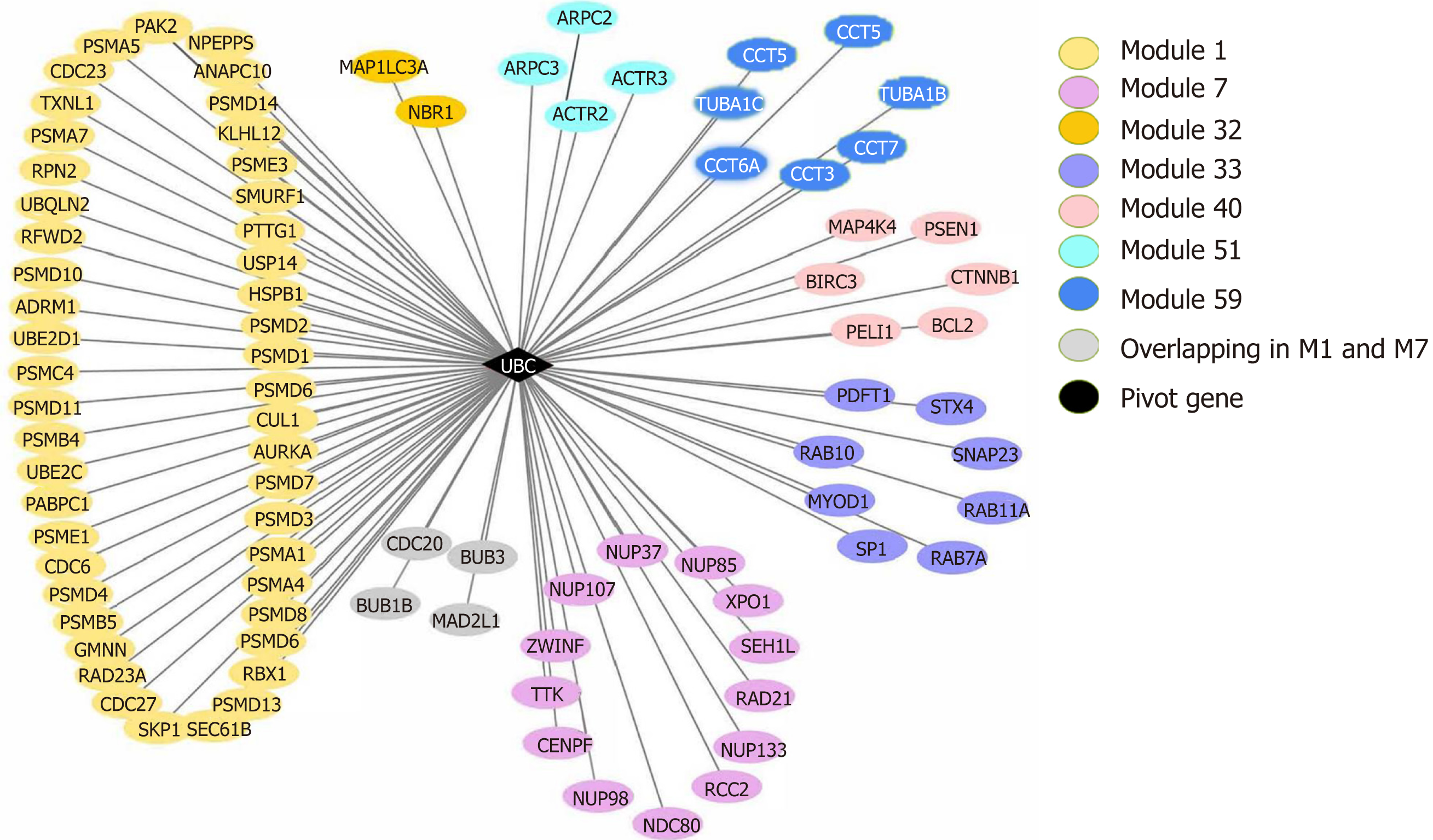
Figure 8 The ubiquitin C gene (pivot) and its connecting modules.
- Citation: Yang YH, Zhang YX, Gui Y, Liu JB, Sun JJ, Fan H. Analysis of the autophagy gene expression profile of pancreatic cancer based on autophagy-related protein microtubule-associated protein 1A/1B-light chain 3. World J Gastroenterol 2019; 25(17): 2086-2098
- URL: https://www.wjgnet.com/1007-9327/full/v25/i17/2086.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i17.2086