Copyright
©The Author(s) 2017.
World J Gastroenterol. Feb 7, 2017; 23(5): 763-775
Published online Feb 7, 2017. doi: 10.3748/wjg.v23.i5.763
Published online Feb 7, 2017. doi: 10.3748/wjg.v23.i5.763
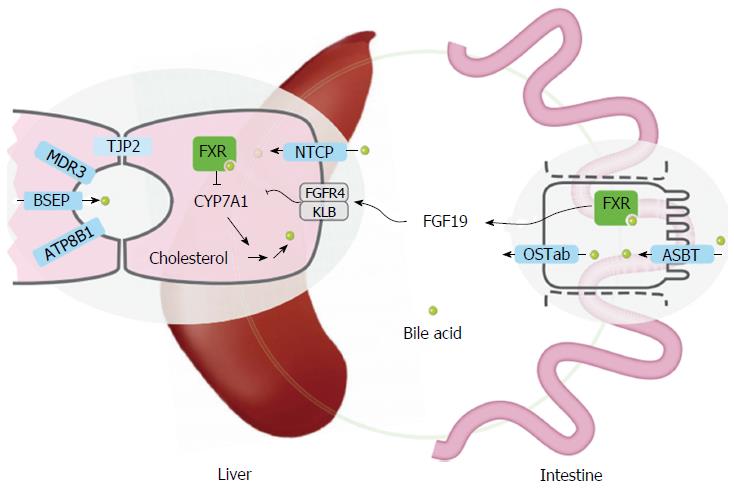
Figure 1 Overview of transporters and sensory proteins involved in hepatic bile flow (regulation).
Inherited cholestatic liver diseases are a consequence of mutations in ATP8B1, ABCB11 (encoding BSEP), ABCB4 (encoding MDR3), or TJP2. These 4 proteins are either expressed in the canalicular membrane or at the tight junctions of the canaliculi (TJP2). NTCP mediates uptake of conjugated bile acids into hepatocytes. ASBT mediates uptake of bile acids from the lumen into the enterocyte and OSTab the bile acid export from the enterocyte into the portal circulation. FXR is a nuclear receptor for bile acids, present in multiple tissues, including liver and intestine. Activation of FXR leads to the enhanced secretion of the hormone FGF19 into the systemic circulation. FGF19 is detected by the heteromeric receptor FGFR4/βKlotho on hepatocytes leading to repression of CYP7A1, and reduced bile acid synthesis. FXR: Farnesoid X receptor; NTCP: Sodium taurocholate cotransporting polypeptide; BSEP: Bile salt export pump; ASBT: Apical sodium-dependent bile acid transporter.
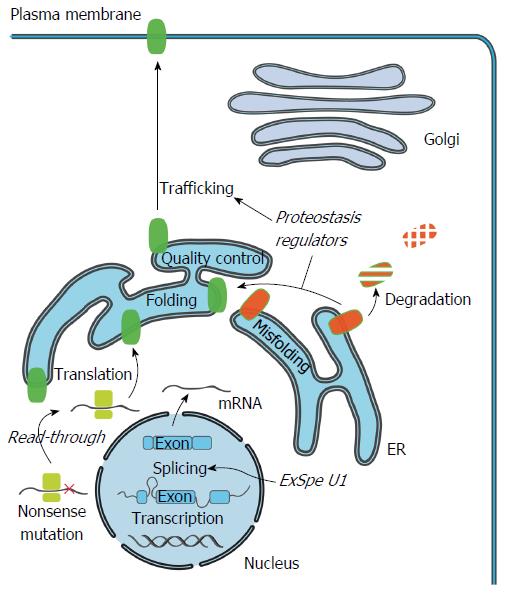
Figure 2 Cellular mechanism of cholestasis-causing mutations and therapeutics.
Specific mutations can impair production of full-length ATP8B1/BSEP/MDR3 protein by induction of premature termination codons (PTC). Compounds such as aminoglycosides and Ataluren, could potentially induce read-through of the PTC and allow translation of full-length transporter protein. Mutations that impair pre-mRNA splicing, protein folding or trafficking can lead to a reduced number or total absence of functional transporters at the cell surface. Pre-mRNA splicing could be restored to enhance the levels of correctly spliced transcripts using modified U1snRNA. Exon specific U1 snRNA (ExSpe U1) provide a special class of modified U1 snRNA molecules that could potentially rescue splicing of multiple mutations in a single exon with less potential side-effects. Misfolded transporters are recognized by the endoplasmic reticulum (ER) quality control system and are targeted for degradation via the ubiquitin-proteasome system. Compounds that modulate the cellular protein homeostasis machinery (proteostasis regulators) can partially rescue the misprocessing, and restore trafficking to the cell surface, largely via unknown mechanisms.
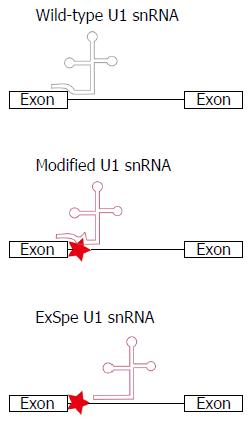
Figure 3 Schematic of the binding of U1 snRNA to exon-intron boundaries to initiate the splicing process.
Disease-associated nucleotide changes can affect the recruitment of the cellular splicing machinery, resulting in splice defects. Modification of the U1 snRNA sequence to restore base-pairing between U1 snRNA and the exon-intron boundary can restore splicing. The use of exon-specific U1 constructs, binding at less conserved intronic regions (ExSpe U1 snRNA), may reduce the risk of off-target effects. Simultaneously, ExSpe U1 snRNAs are likely to be effective in a higher number of patients as they can restore splice defects due to several distinct mutations within a certain exon-intron boundary.
- Citation: van der Woerd WL, Houwen RH, van de Graaf SF. Current and future therapies for inherited cholestatic liver diseases. World J Gastroenterol 2017; 23(5): 763-775
- URL: https://www.wjgnet.com/1007-9327/full/v23/i5/763.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i5.763