Copyright
©The Author(s) 2017.
World J Gastroenterol. Sep 14, 2017; 23(34): 6330-6338
Published online Sep 14, 2017. doi: 10.3748/wjg.v23.i34.6330
Published online Sep 14, 2017. doi: 10.3748/wjg.v23.i34.6330
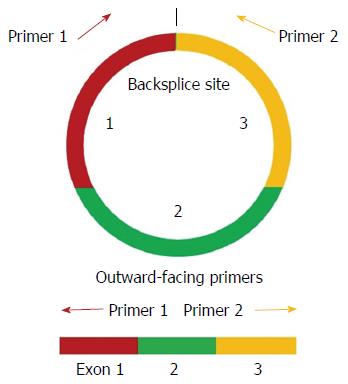
Figure 1 Schematic diagram of the polymerase chain reaction primers used to specifically detect circular transcripts.
Divergent polymerase chain reaction primers, rather than the more commonly used convergent primers, were designed for the circRNAs.
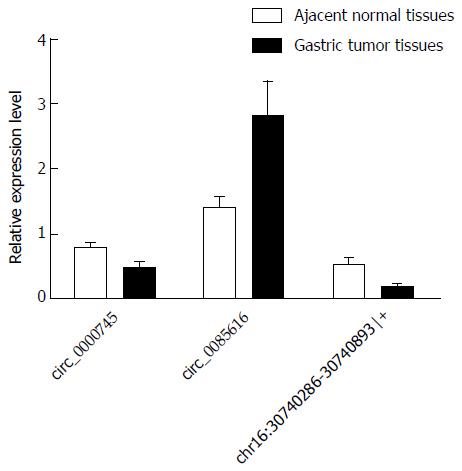
Figure 2 Validation of the expression levels of the three circRNAs using quantitative reverse transcription-polymerase chain reaction analysis.
The three differently expressed circRNAs were validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in 20 gastric tumor tissues and paired adjacent non-tumor tissues. The qRT-PCR results are presented as mean and SE of the mean.
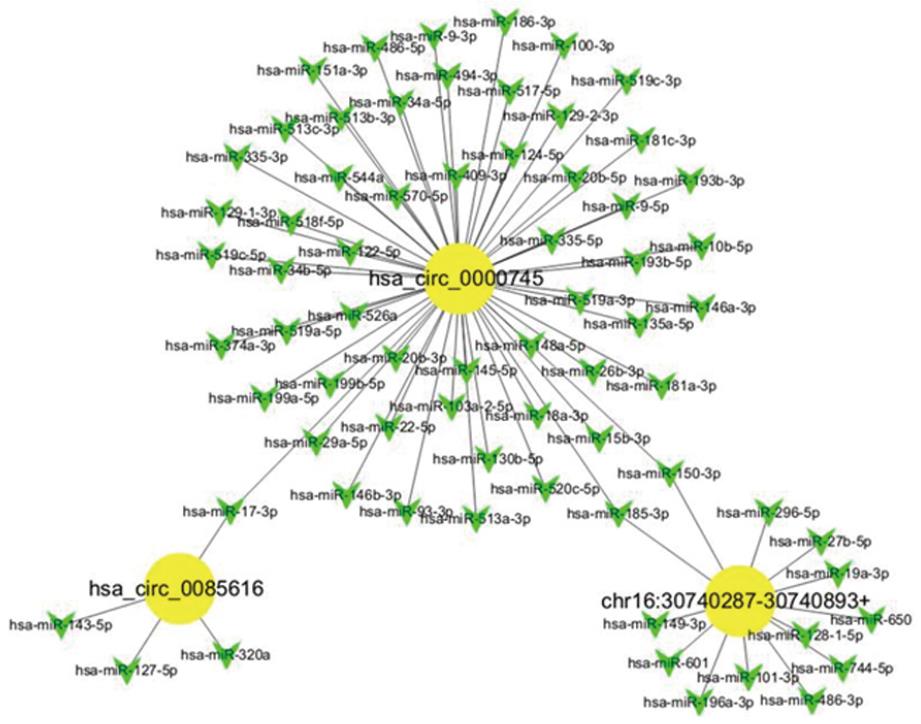
Figure 3 Network of circular RNAs and the predicted binding miRNAs.
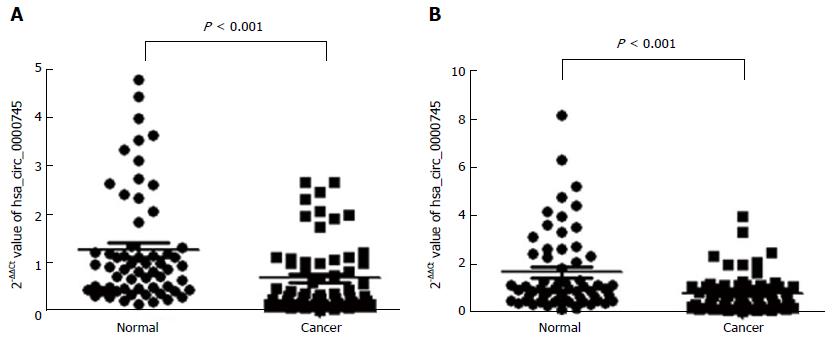
Figure 4 Hsa_circ_0000745 expression levels.
A: Hsa_circ_0000745 expression levels in gastric cancer tissues and paired adjacent non-tumor tissues; B: Hsa_circ_0000745 expression levels in plasma samples from patients with gastric cancer and healthy volunteers. P < 0.001, n = 60. The expression is shown as the 2- ΔΔCt value.
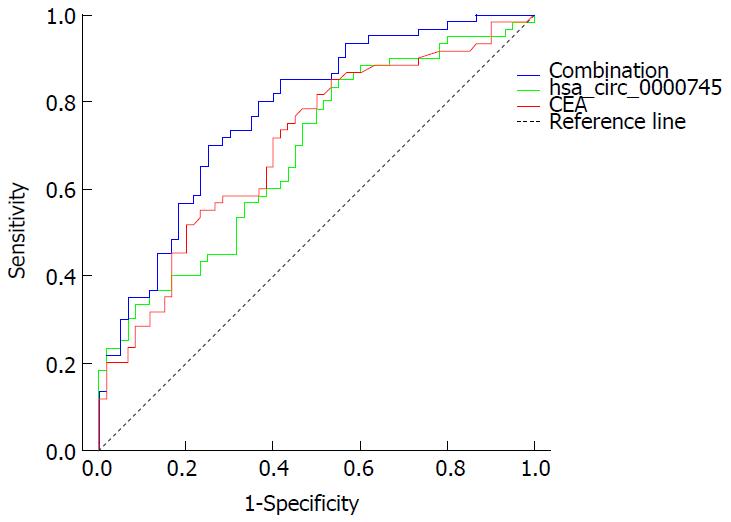
Figure 5 Receiver operating characteristic curve of plasma hsa_circ_0000745 and serum carcinoembryogenic antigen levels.
CEA: Carcinoembryogenic antigen.
- Citation: Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol 2017; 23(34): 6330-6338
- URL: https://www.wjgnet.com/1007-9327/full/v23/i34/6330.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i34.6330