Copyright
©The Author(s) 2016.
World J Gastroenterol. Feb 21, 2016; 22(7): 2271-2283
Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2271
Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2271
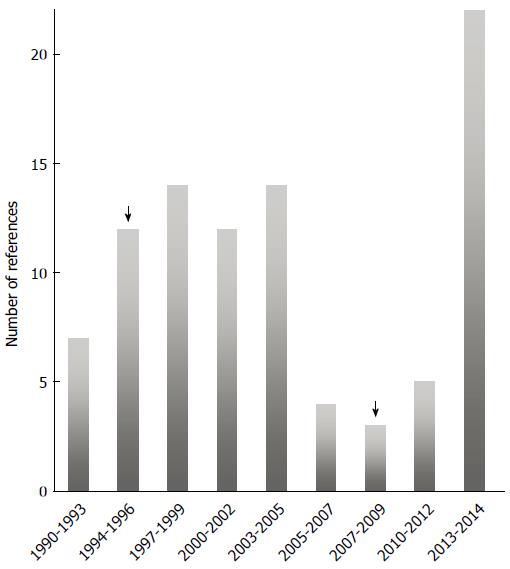
Figure 1 Timeline of the number of papers about hepatitis E virus.
Based on references found in PubMed when hepatitis E virus (HEV) was used as a search keyword. All the years prior to 1990 clustered at first time point. Awareness was more prominent following the demonstration of chronic hepatitis as caused by HEV. Arrows highlights first report of zoonotic transmission (left arrow), and the demonstration of chronicity due to HEV infection (right arrow).
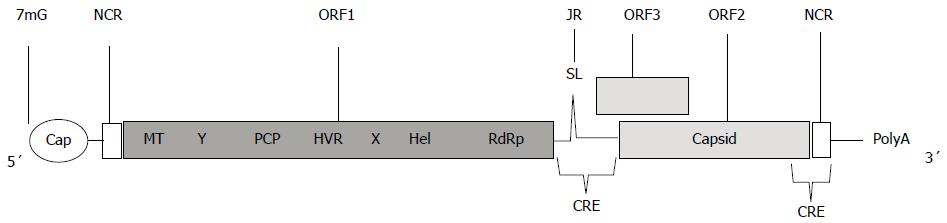
Figure 2 A schematic representation of the RNA- hepatitis E virus genome.
RNA encodes for multiple proteins from three Open Reading Frames (ORFs). ORF1 comprises the methyltransferase (MT), Y domain, a papain-like cysteine protease (PCP), hypervariable region (HVR), X macrodomain, helicase (Hel), and RNA-dependent RNA polymerase (RdRp). ORF 2 encodes the capsid protein, and ORF3 encodes a cytoskeleton-associated phosphoprotein. 5´Cap and 3´PolyA modifications are also depicted. The junction region (JR), stem-loop structure (SL), and cis-reactive element (CRE) have been described elsewhere[12].
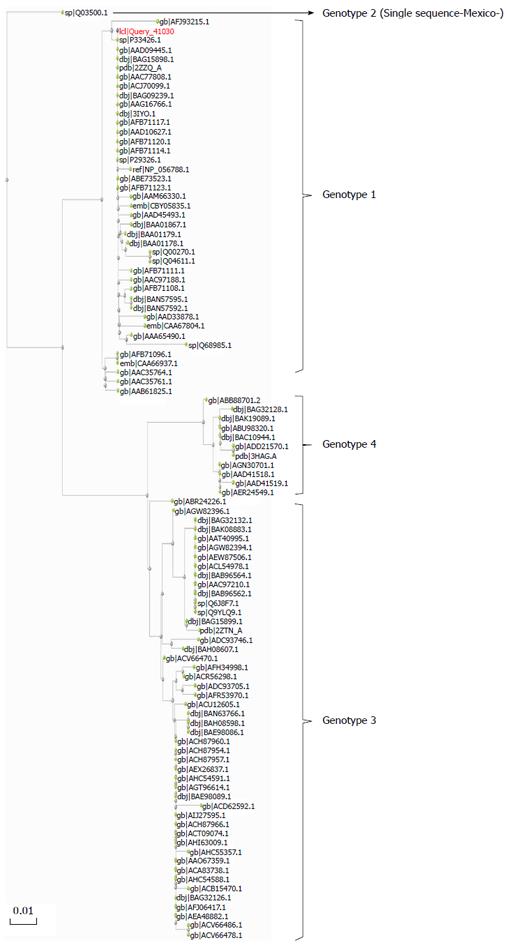
Figure 3 A phylogenetic tree for hepatitis E virus according to their amino-acid sequences from the recognition domain E2 (E domain, residues 455 to 606, based on US-1 strain) in the open reading frame capsid protein.
This region corresponds with the binding domain of neutralizing Mab 8C11[32,33]. It contains all epitopes demonstrated to elicit neutralizing antibodies. This region is sufficient to provide protection, and is the basis of the current recombinant vaccine Hecolin 293 approved in China. The analysis was based on 100 sequences representing the diversity of HEV sequences. The tree was constructed by the Maximum Likelihood method using PhyML software, as implemented in server http://phylogeny.lirmm.fr[33]. The scale bar represents amino-acid substitutions and the statistical support was obtained from 100 Bootstrap replicates.
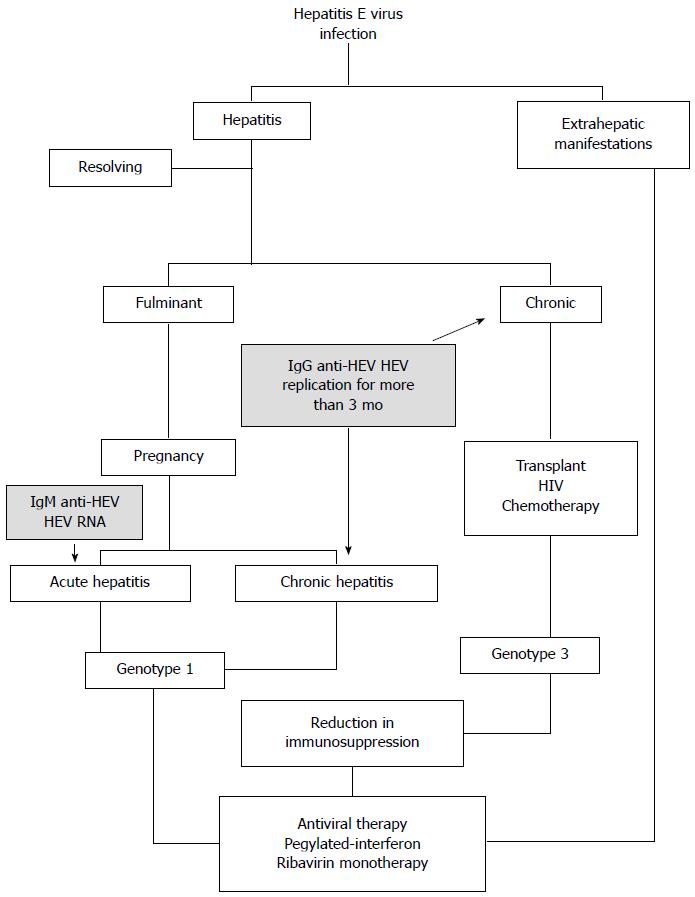
Figure 4 Patterns of hepatitis E virus infection.
The typical clinical courses taken by hepatitis E virus (HEV) infections are depicted. The scheme is based on the clinical approach followed at different institutions, as first revised in Kamar et al[13]. The shaded text corresponds to the clinical laboratory markers used to classify the degree of disease caused. This model is based mainly on infections with HEV from genotypes 1 and 3, as shown. Fewer data are available for genotypes 2 and 4, but the same general model may apply.
- Citation: Fierro NA, Realpe M, Meraz-Medina T, Roman S, Panduro A. Hepatitis E virus: An ancient hidden enemy in Latin America. World J Gastroenterol 2016; 22(7): 2271-2283
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2271.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2271