Copyright
©The Author(s) 2016.
World J Gastroenterol. Jul 14, 2016; 22(26): 5971-6007
Published online Jul 14, 2016. doi: 10.3748/wjg.v22.i26.5971
Published online Jul 14, 2016. doi: 10.3748/wjg.v22.i26.5971
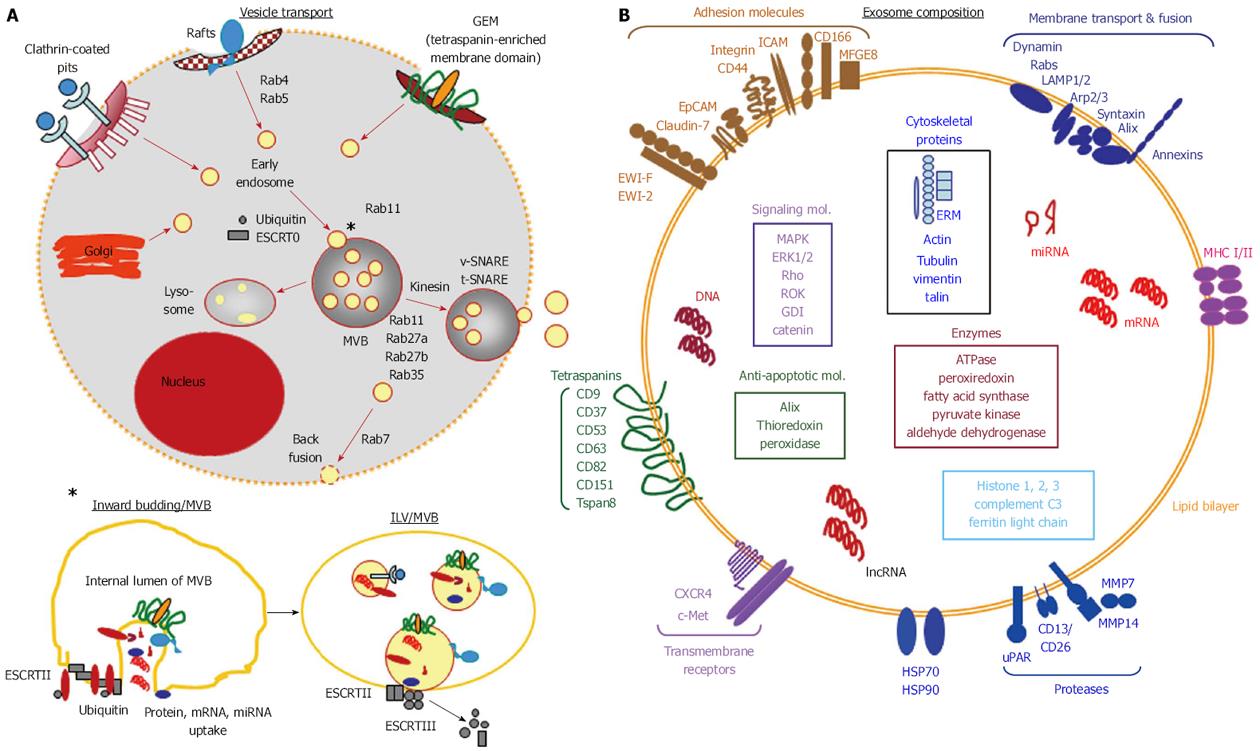
Figure 1 Exosome biogenesis and release.
A: Exosome biogenesis is initiated by the generation of early endosomes delivered by the Golgi complex or by invagination of defined membrane microdomains such as clathrin coated pits, rafts and glycolipid enriched membrane microdomains, which are by scission separated from the originating membrane. Early endosomes are budding into multivesicular bodies During this process the small cytoplasm of endosomes is loaded with proteins, mRNA and DNA. The invaginated early endosomes dissociate from the membrane of multivesicular bodies and are then called intraluminal vesicles. Early endosomes and multivesicular bodies are guided along microtubuli by transporter proteins and transporter complexes, where monoubiquitination, different Rabs, ESCRT complexes and SNARE proteins account for distinct guiding routes; B: Exosomes are characterized by a lipid bilayer enriched in cholesterol, sphingomyelin, GM3 and phosphatidylserine and membrane proteins that vary with the donor cell and the originating membrane microdomain, tetraspanins being a constitutive and highly enriched component. Signal transduction and cytoskeletal proteins are mostly recruited via the association with the inner plasma membrane. Cytosolic proteins, coding and noncoding RNA and DNA are selectively recruited. Of note, to our knowledge, all CSC markers are recovered and enriched in exosomes. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; ESCRT: Endosomal sorting complex required for transport; MVB: Multivesicular bodies; ILV: Intraluminal vesicles.
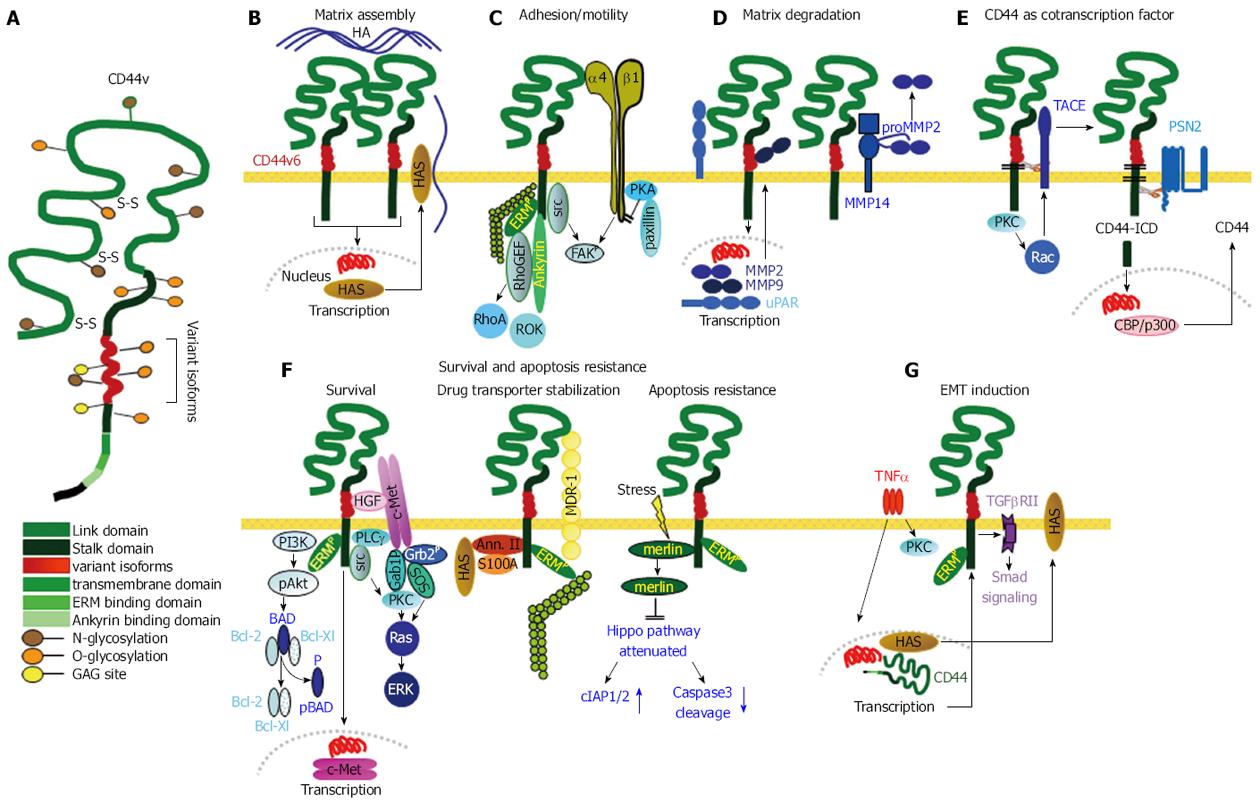
Figure 2 Cancer stem cells supporting activities of CD44v6.
A: Structure of CD44 including insertion of variant exon products; B: CD44v6 is engaged in assembling the HA matrix by inducing HAS transcription. The matrix supports CD44 activation; C: GEM-located CD44/CD44v6 is src associated and cooperates with adjacent integrins. Activated ERM proteins and Ankyrin link CD44 to the cytoskeleton and initiate RhoA and ROK activation, which together play an important role in CSC motility; D: CD44v6 is engaged in uPAR and MMP transcription. It associates with MMP14, which captures MMP2 and MMP9, supporting ECM degradation and remodeling; E: GEM-located CD44v6 can be cleaved by TACE and subsequently by PSN2. CD44-ICD is a powerful cotranscription factor, which induces beside others CD44 transcription; F: There are several pathways, whereby CD44v6 supports CSC survival and apoptosis resistance. It cooperates with c-Met, which is brought into vicinity by CD44v6-bound HGF; a complex of CD44, Annexin II, S100A and HAS stabilizes MDR1; stress induces merlin phosphorylation, which dissociates from CD44 and hampers activation of the Hippo pathway; G: One pathway of CD44-linked induction of EMT relies on TNFα that fosters CD44 expression and CD44 cooperativity with TGFβRII promoting Smad signaling. CD44 also is engaged in activation of Nanog and in repressing miRNA transcription that targets EMT proteins (not shown). The majority of these CSC supporting activities of CD44/CD44v6 rely on GEM-recruited CD44. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; ERM: Cytoskeletal proteins ezrin, radixin, moesin; EMT: Epithelial mesenchymal transition; TNFα: Tumor necrosis factor α; TGFβ: Transforming growth factor β; TACE: TNFα converting enzyme.
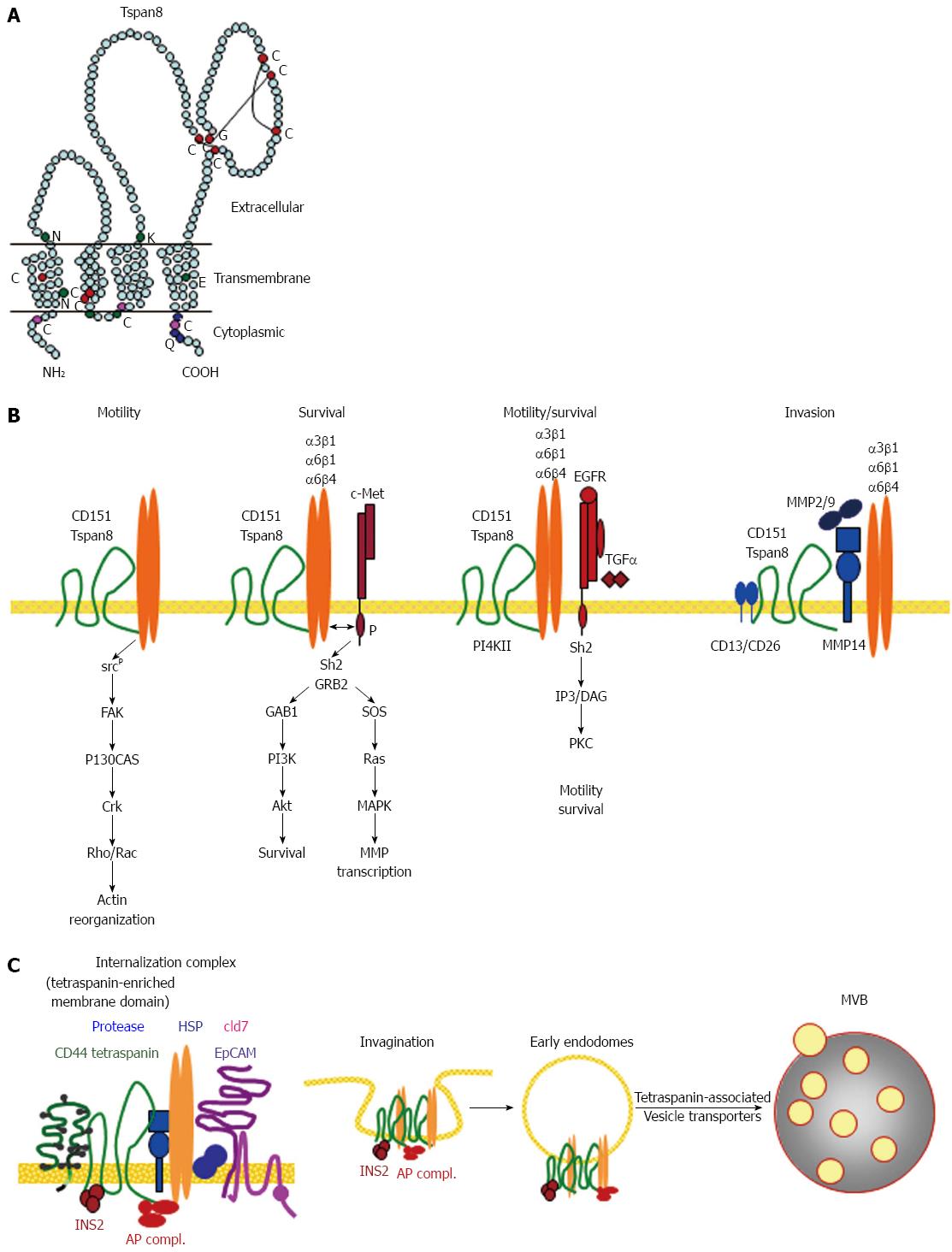
Figure 3 Cancer stem cells supporting activities of Tspan8.
A: The structure of Tspan8 showing the prominent large extracellular loop, which is the main binding site for laterally associated molecules; B: Dominant partners of Tspan8 and CD151 are integrins and proteases. Tspan8 and CD151 also associate with RTK, CD44v6 and EpCAM. The integrin associations promote CSC motility, the RTK associations survival and the protease associations invasiveness: C: the strongest contribution of Tspan8 in support of CSC relies on its engagement in GEM complex located molecule internalization. Tspan8 is also involved in vesicle traffic. The importance of Tspan8 in CSC is linked to recruiting additional CSC markers into GEM and in contributing to the transfer of the GEM complex into TEX. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; MVB: Multivesicular bodies; PKC: Phophokinase C; RTK: Receptor tyrosine kinase.
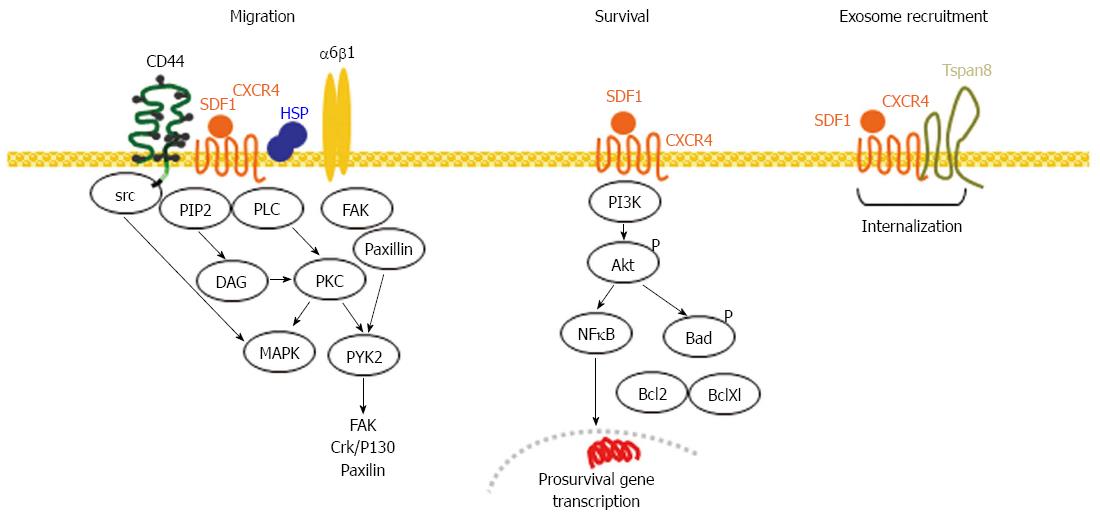
Figure 4 CXCR4, a marker of migrating cancer stem cells.
CXCR4 becomes activated by SDF1 binding, which in concert with CD44, integrins and HSP initiates several signaling cascades that promote directed motility. CXCR4 also activates the PI3K/Akt pathway promoting antiapoptotic protein activation and prosurvival gene transcription. Activated CXCR4 becomes recruited into GEM, where in Pa-CSC the association with Tspan8 supports internalization. CXCR4 is recovered in TEX. In Pa-CSC CXCR4 cooperates with CD44(v6), laminin binding integrins and Tspan8 in TEM and is recruited into TEX. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; NFκB: Nuclear factor κB; TEX: Tumor exosomes; HSP: Heat shock protein; PLC: Phospholipase C; PKC: Phophokinase C; PI3K: Phosphatidylinositol-4,5-bophosphate 3 kinase; MAPK: Mitogen-activazed protein kinase; SDF1: Stroma-derived factor 1.
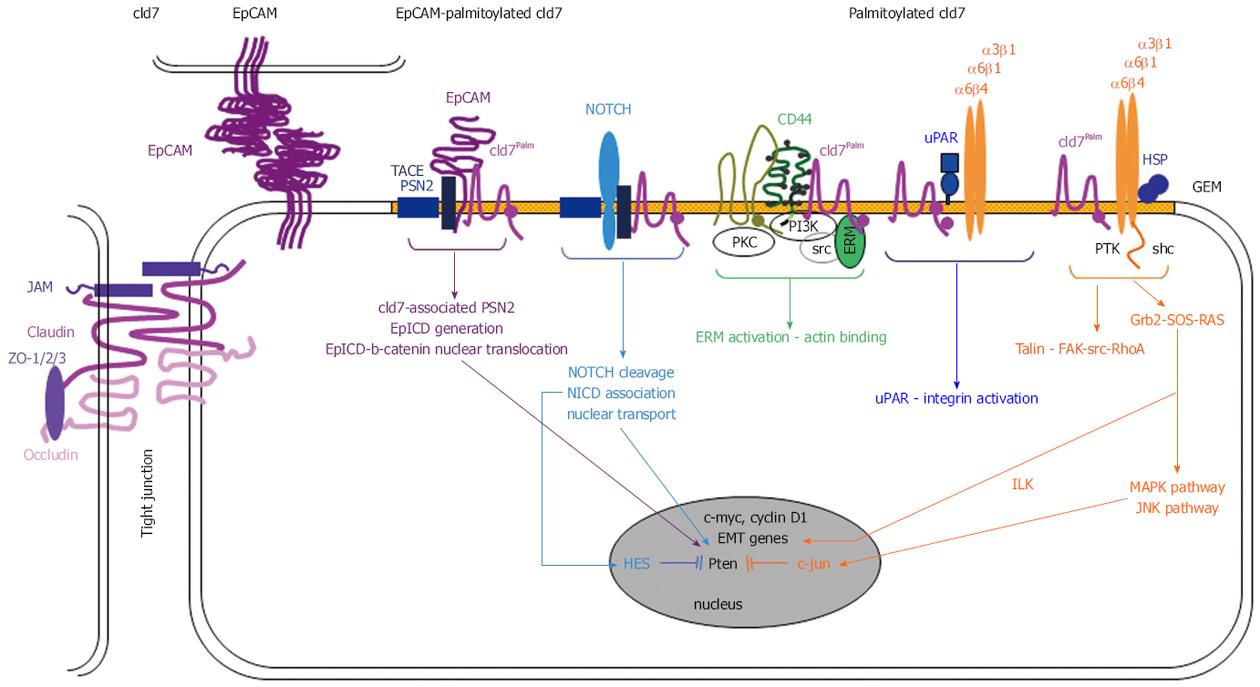
Figure 5 The EpCAM-palmitoylated claudin7 complex in pancreatic cancer stem cells.
Upon palmitoylation the TJ protein cld7 becomes excluded from TJ and recruited into GEM, where it associates with monomeric EpC. Monomeric EpC becomes susceptible to GEM-located TACE and PSN2. EpICD associates with β-catenin and translocates to the nucleus acting as a cotranscription factor for c-myc, cyclinD1 and several EMT genes. The cld7-TACE-PSN2 complex also contributes to NOTCH cleavage. NICD contributes to HES activation, which inhibits Pten transcription. The palmitoylated cld7-uPAR association promotes integrin activation and transcription of EMT genes via ILK. Activation of the JNK pathway contributes to inhibition of Pten transcription. In Pa-CSC EpC becomes cleaved and involved in EMT gene transcription; palmitoylated cld7 additionally is engaged via Notch cleavage and integrin activation in apoptosis resistance. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; ILK: integrin-linked kinase; TJ: Tight junction; HSP: Heat shock protein; PKC: Phophokinase C; TACE: TNFα converting enzyme.
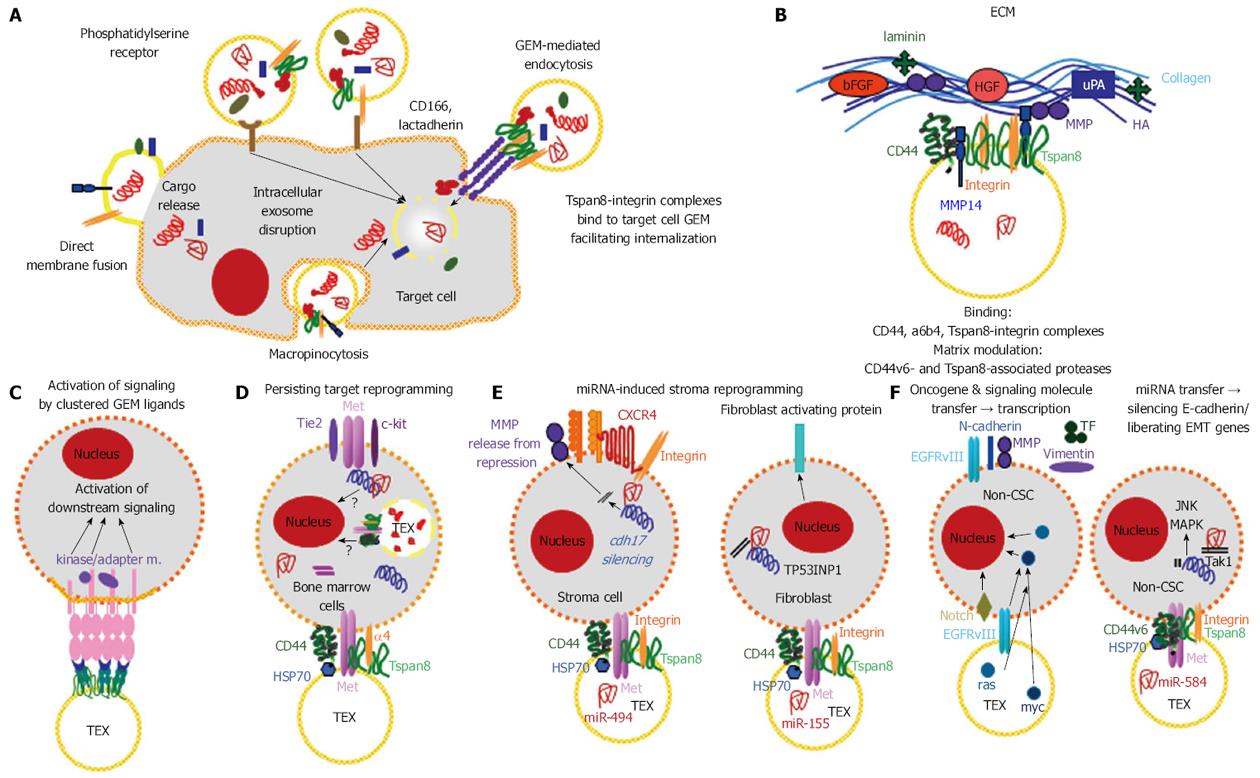
Figure 6 Contribution of exosomal cancer stem cell markers to target selection, binding, uptake and target modulation.
A: Exosomes uptake by cells can proceed via membrane fusion, macropinocytosis, receptor ligand binding. GEM-derived exosomes bind to complexes of ligands located in internalization prone membrane microdomains, which increases selectivity of uptake and facilitates uptake; B: Binding to the ECM is facilitated by the Pa-CSC markers α6β4 and CD44v6. CD44v6- and Tspan8-associated proteases facilitate matrix degradation; C-E: There are multiple pathways whereby TEX affect target cells, only selected examples are shown: Signal transduction can be initiated by clustering GEM ligands; in progenitor cells, e.g., in the bone marrow, activation of signaling molecules can initiate a shift towards differentiation; the transfer of TEX miRNA can initiate release from repression by the miRNA target or tumor suppressor RNA can become silenced such that resting mesenchymal cells turn into an activated phenotype; F: EMT was induced in Non-CSC by the transfer of oncogenes, activation of EMT-related transcription factors or miRNA blocking transcription of RNA engaged in epithelial stage maintenance. CSC: Cancer stem cells; GEM: Glycolipid-enriched microdomains; ECM: Extracellular matrix; EMT: Epithelial mesenchymal transition; HA: Hyaluronan; TEX: Tumor exosomes.
- Citation: Heiler S, Wang Z, Zöller M. Pancreatic cancer stem cell markers and exosomes - the incentive push. World J Gastroenterol 2016; 22(26): 5971-6007
- URL: https://www.wjgnet.com/1007-9327/full/v22/i26/5971.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i26.5971