Copyright
©The Author(s) 2015.
World J Gastroenterol. Jan 7, 2015; 21(1): 84-93
Published online Jan 7, 2015. doi: 10.3748/wjg.v21.i1.84
Published online Jan 7, 2015. doi: 10.3748/wjg.v21.i1.84
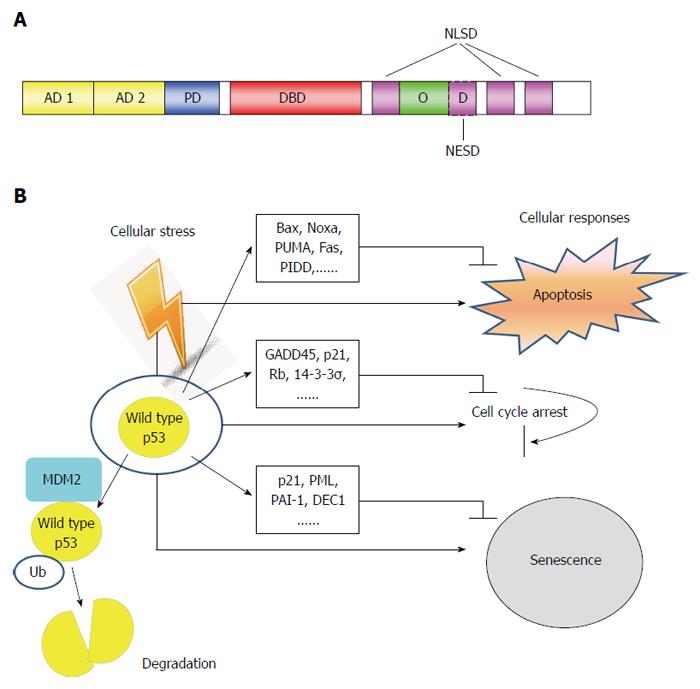
Figure 1 Structure and function of p53 tumor suppressor.
A: Schematic of p53 protein structure. The function domains and corresponding amino acid regions are indicated. N-terminus transcription-activation domain (TAD): Residues 1-63; AD1: Residues 1-42 for G1 arrest and apoptotic activity; AD2: Residues 43-63 important for senescence-activity; PD: Residues 64-92 important for apoptotic activity; DBD: Residues 102-292 responsible for binding the p53 co-repressors; NLSD: Residues 316-324, 370-376, 380-386; OD: Residues 325-356; NESD: Residues 340-353; B: In normal cells, p53 activates a plethora of target genes involved in diverse biological processes in response to cellular stress. Ub: Ubiquitin; PD: Poly-proline domain; DBD: DNA binding core domain; OD: Homo-oligomerization domain; NESD: Nuclear export signaling domain; NLSD: Nuclear localization signaling domain.
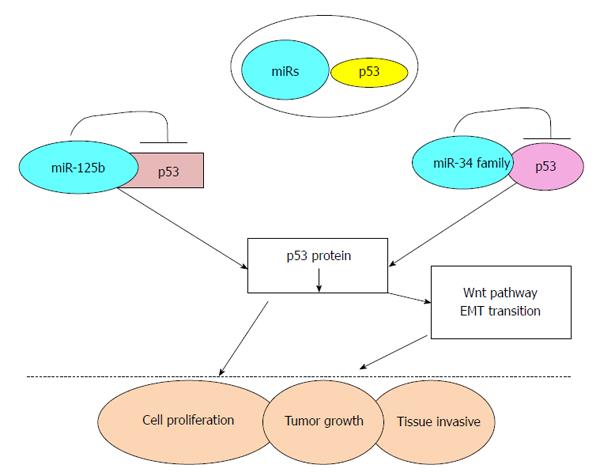
Figure 2 Schematic representation of miRNAs regulating p53 pathway and subsequent tumorigenesis.
Figure 3 Small molecule compounds pharmacologically reactivating of p53 function.
MI43 and Nutlin-3 bound to MDM2 blocking MDM2-p53 interaction. RITA bound to p53 interfering MDM2-p53 interaction. α-Lipoic acid increased p53 protein stability and its apoptotic effect. Quinacrine induced the autophagy-associated cell death in a p53-dependent manner. NSC17632 activated p53-like activatity dependent on p73. PRIMA-1/PRIMA-1MET restored mutant p53 to exert apoptotic effect. Maslinic acid and Epicatechin gallate as plant extraction modulated the expression of p53 and its target genes in p53-dependent apoptotic and cell cycle arrest pathway.
-
Citation: Li XL, Zhou J, Chen ZR, Chng WJ.
p53 mutations in colorectal cancer- molecular pathogenesis and pharmacological reactivation. World J Gastroenterol 2015; 21(1): 84-93 - URL: https://www.wjgnet.com/1007-9327/full/v21/i1/84.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i1.84