Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 21, 2014; 20(3): 795-803
Published online Jan 21, 2014. doi: 10.3748/wjg.v20.i3.795
Published online Jan 21, 2014. doi: 10.3748/wjg.v20.i3.795
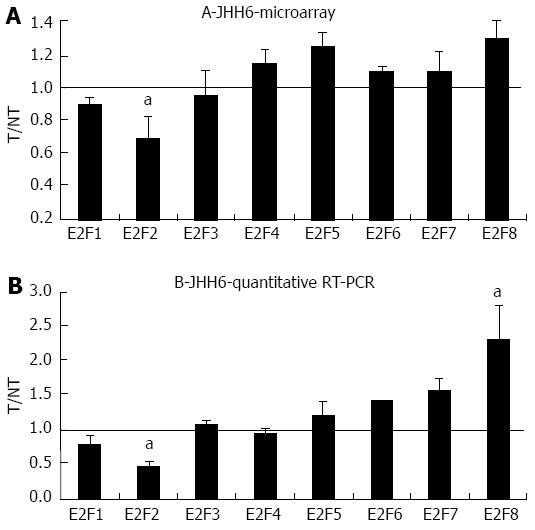
Figure 1 Microarray and quantitative real-time reverse transcription polymerase chain reaction assays in JHH6 following bortezomib treatment.
A: The mRNA levels of the indicated E2Fs were evaluated by microarray analysis two days after JHH6 treatment with 40 nmol/L bortezomib. Data are reported as ratio between treated cells (T) and non-treated cells (NT); data are shown as means ± SEM; aP < 0.05 vs controls; depending on the E2F member the number of evaluation ranged from three up to ten; B: The mRNA levels of the indicated E2Fs were evaluated by quantitative real-time reverse transcription polymerase chain reaction assay; data, normalized to GAPDH mRNA, are reported as ratio between T and NT; data are reported as means ± SEM; aP < 0.05 vs controls, n = 5. RT-PCR: Reverse transcription polymerase chain reaction.
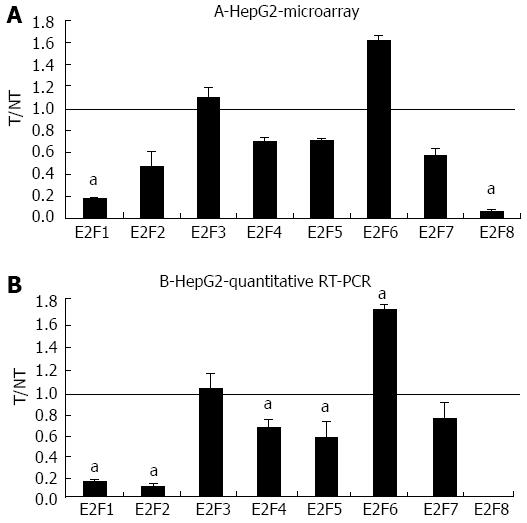
Figure 2 Microarray and quantitative real-time reverse transcription polymerase chain reaction assays in HepG2 following bortezomib treatment.
A: The mRNA levels of the indicated E2Fs were evaluated by microarray analysis two days after JHH6 treatment with 40 nmol/L bortezomib. Data are reported as ratio between treated cells (T) and non-treated cells (NT); data are shown as means ± SEM aP < 0.05 vs controls; depending on the E2F member the number of evaluation ranged from three up to ten. B: The mRNA levels of the indicated E2Fs were evaluated by quantitative real-time reverse transcription polymerase chain assay; data, normalized to GAPDH mRNA, are shown as ratio between T and NT; data are reported as means ± SEM; aP < 0.05 vs controls, n = 5. RT-PCR: Reverse transcription polymerase chain reaction.
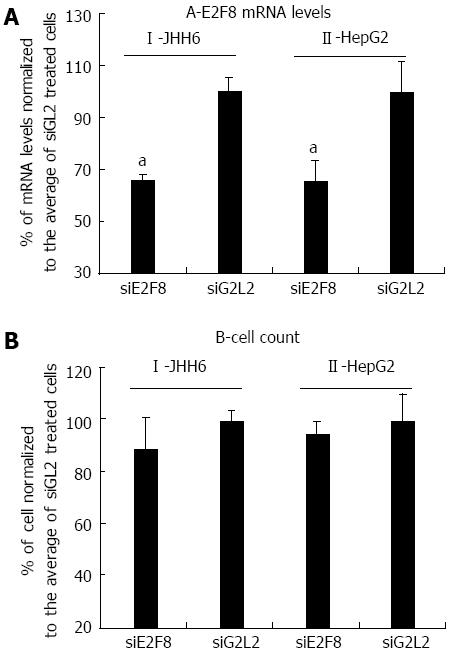
Figure 3 Effects of E2F8 depletion by siRNA in HepG2 and JHH6.
A: Four and 2 d after siRNA administration, the reductions of E2F8 mRNA in JHH6 and HepG2, respectively, are reported; the data, normalized to GAPDH levels, are shown as mean ± SEM; aP < 0.05 vs controls, n = 6; B: the effects of siE2F8 on cell number 4 and 2 d after siRNA administration in JHH6 and HepG2, respectively, are reported; the data, normalized to siGL2 treated cells, are shown as mean ± SEM, n = 6. siE2F8: siRNA against E2F8 mRNA; siGL2: Control siRNA against the luciferase mRNA.
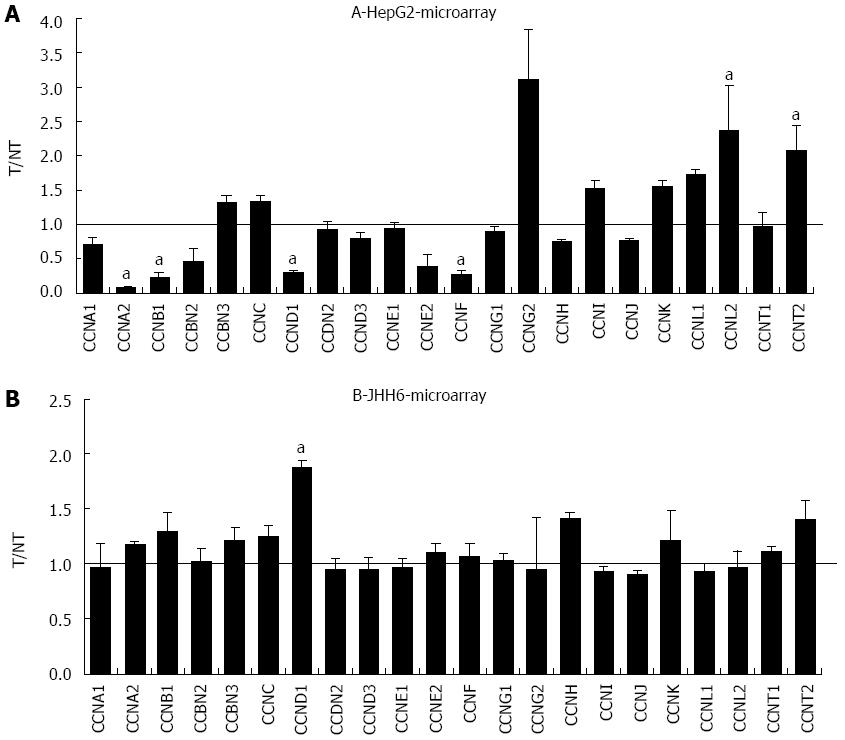
Figure 4 Microarray assay in HepG2 and JHH6 following Bortezomib treatment.
The mRNA levels of the indicated cyclins were evaluated by microarray analysis two days after HepG2 (A) or JHH6 (B) treatment by 40 nmol/L Bortezomib. Data are reported as ratio between treated cells (T) and non-treated cells (NT); data are shown as means ± SEM; aP < 0.05 vs controls; depending on the cyclin member, the number of microarray evaluation ranged from three up to ten.
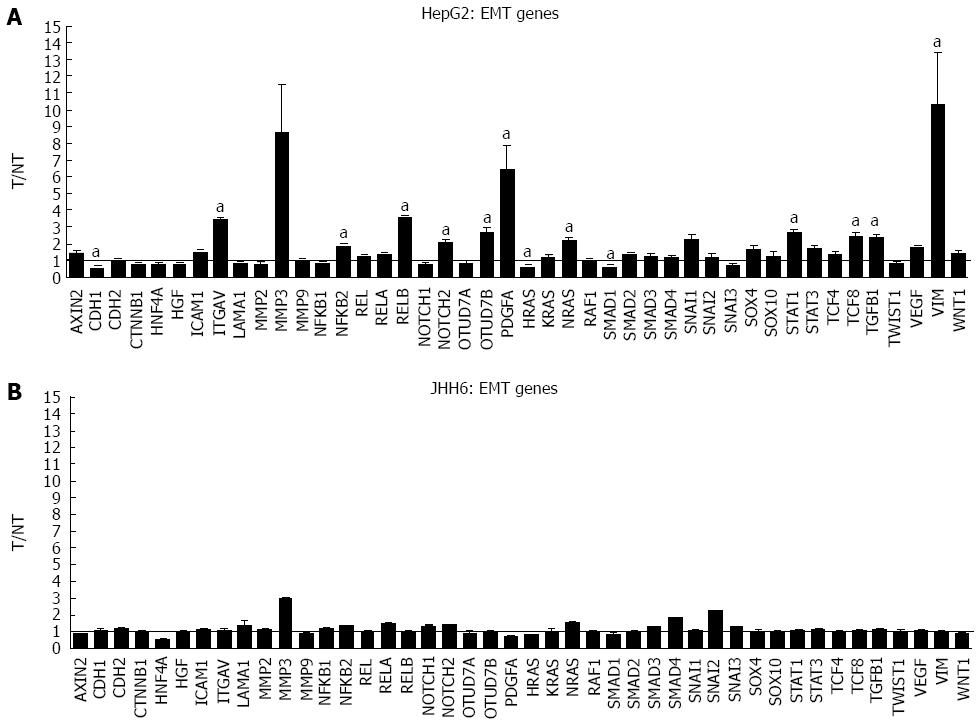
Figure 5 Microarray assays in HepG2 and JHH6 following bortezomib treatment.
The mRNA levels of the indicated epithelial-mesenchymal transition (EMT) genes were evaluated by microarray analysis two days after HepG2 (A) or JHH6 (B) treatment by 40 nmol/L Bortezomib. Data are reported as ratio between treated cells (T) and non-treated cells (NT); data are shown as means ± SEM; aP < 0.05 vs controls; depending on the EMT member, the number of microarray evaluation ranged from three up to ten.
- Citation: Baiz D, Dapas B, Farra R, Scaggiante B, Pozzato G, Zanconati F, Fiotti N, Consoloni L, Chiaretti S, Grassi G. Bortezomib effect on E2F and cyclin family members in human hepatocellular carcinoma cell lines. World J Gastroenterol 2014; 20(3): 795-803
- URL: https://www.wjgnet.com/1007-9327/full/v20/i3/795.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i3.795