Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 14, 2014; 20(2): 425-435
Published online Jan 14, 2014. doi: 10.3748/wjg.v20.i2.425
Published online Jan 14, 2014. doi: 10.3748/wjg.v20.i2.425
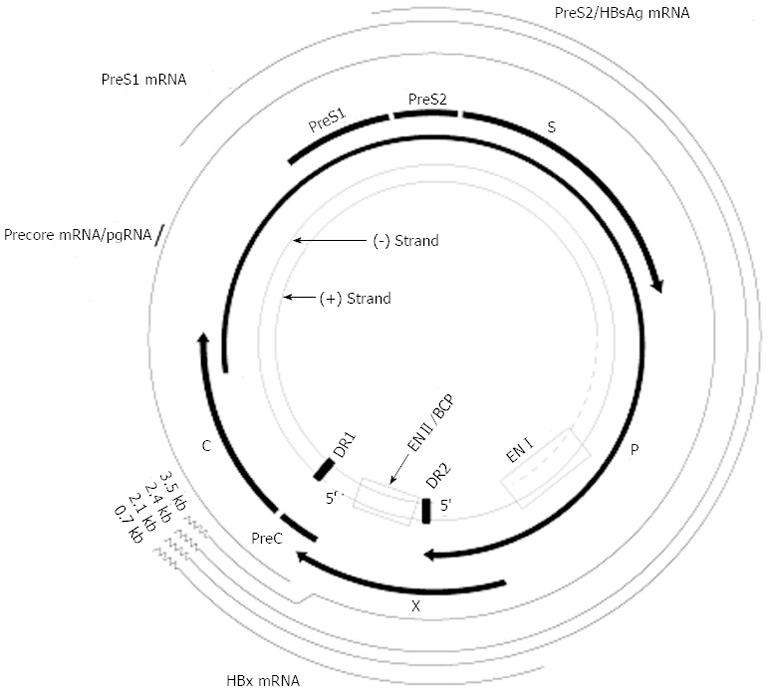
Figure 1 Genomic organization of hepatitis B virus.
The inner circle depicts the rcDNA including the complete minus-strand DNA and the incomplete plus-strand DNA. The direct repeats, DR1 and DR2, as well as the two enhancers, ENI and ENII, are shown. The outer circle depicts the four viral RNAs, the core (C) or pgRNA, the pre-S (L) mRNA, the S mRNA, and the X mRNA. The common 3’-ends (poly-A) of the three mRNAs are indicated by the curve lines. The four protein-coding regions are shown including the precore (PC) and core genes, the polymerase gene, and the X gene. The envelope genes pre-S1 (L), pre-S2 (M), and surface (S) overlap with the polymerase open reading frame.
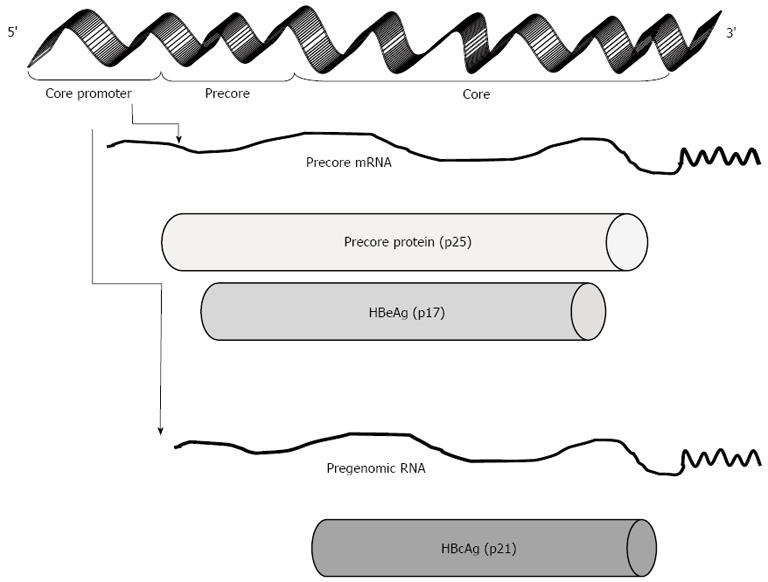
Figure 2 Transcription and translation of hepatitis B virus precore.
The core promoter region regulates the transcription of precore mRNA and pregenomic RNA from the same open reading frame. The precore mRNA is translated into precore protein, which is processed at the N-terminal and C-terminal ends to HBeAg, a secretory protein. Pregenomic RNA is reverse transcribed into HBV DNA and also translated into core protein (HBcAg), which overlaps with HBeAg. HBcAg: Hepatitis B core protein antigen; HBeAg: Hepatitis B e antigen.
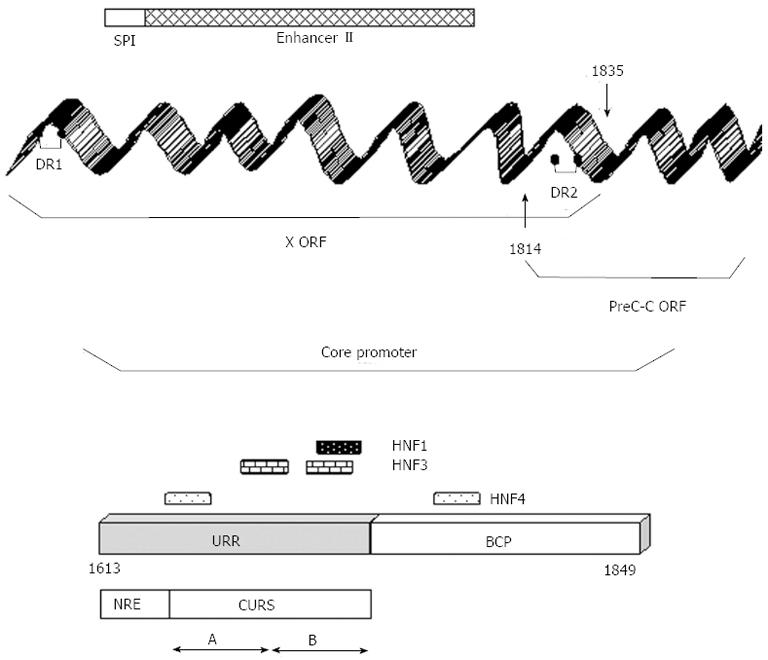
Figure 3 Molecular biology of the hepatitis B virus core promoter.
The core promoter (CP) overlaps the 3’-end of the X open reading frame (ORF) and the 5’-end of the pre-C/C ORF. The CP comprises the upper regulatory region (URR) and the basic core promoter (BCP). The former involves a negative regulatory element (NRE) and a core upstream regulatory sequence (CURS) which can be further subdivided into two domains: CURS-A and CURS-B. The sequence of enhancer II and binding sites hepatocyte nuclear factor 1, 3 and 4 (HNF1, HNF3, HNF4) are shown. SPI: S promoter; DR: direct repeat.
- Citation: Quarleri J. Core promoter: A critical region where the hepatitis B virus makes decisions. World J Gastroenterol 2014; 20(2): 425-435
- URL: https://www.wjgnet.com/1007-9327/full/v20/i2/425.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i2.425