Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Jul 21, 2013; 19(27): 4289-4299
Published online Jul 21, 2013. doi: 10.3748/wjg.v19.i27.4289
Published online Jul 21, 2013. doi: 10.3748/wjg.v19.i27.4289
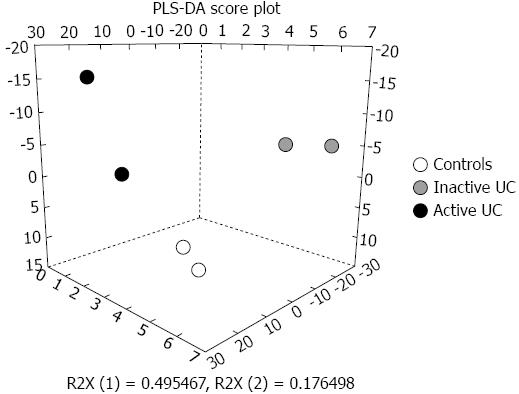
Figure 1 Projection to latent structure-discriminant analysis score-plot of the miRNA microarray expression profiles from mucosal colonic biopsies of controls, active ulcerative colitis, and inactive ulcerative colitis reveals a clear separation of these three groups.
All patient with active ulcerative colitis (UC) are positioned in the left part of the space, and all patients with inactive UC are placed in the right space, whereas all control subjects are found in the middle. PLS-DA: Projection to latent structure-discriminant analysis.
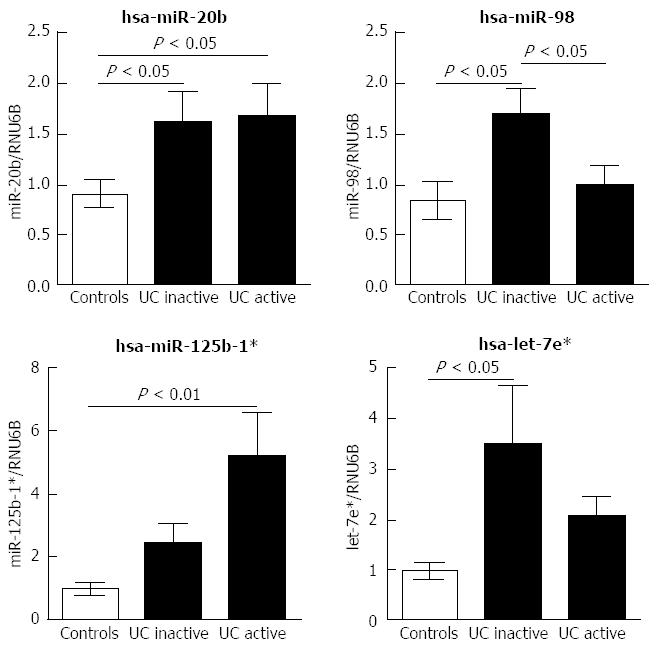
Figure 2 Expression levels of four miRNAs in mucosal colonic biopsies were significantly up-regulated in ulcerative colitis.
Expression differences of miRNA in active ulcerative colitis (UC) (n = 20), inactive UC (n = 19), and controls (n = 20). Levels were determined using quantitative real-time polymerase chain reaction. Relative expression differences of each miRNA were normalized to endogenous RNU6B expression and calculated using the 2-∆∆CT method. P value was calculated by Mann-Whitney U test and data are represented as medians with interquartile ranges.
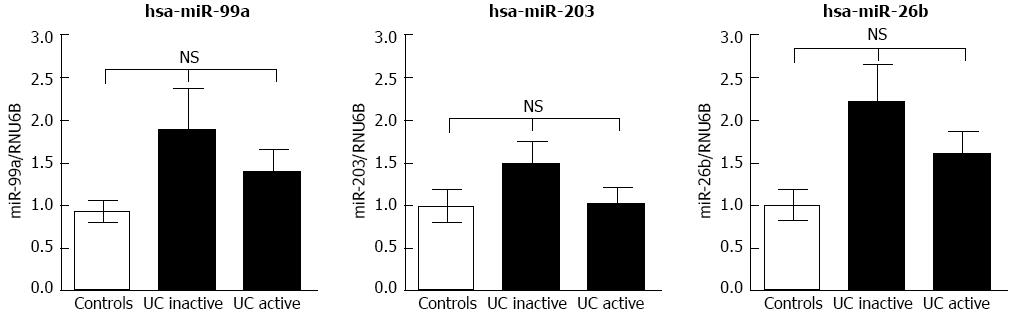
Figure 3 Expression of other miRNAs identified by microarray profiling.
Expression differences of miRNA in active ulcerative colitis (UC) (n = 20), inactive UC (n = 19), and controls (n = 20). Levels were determined using quantitative real-time polymerase chain reaction. Relative expression differences of each miRNA were normalized to endogenous RNU6B expression and calculated using the 2-∆∆CT method. NS: Not significant.
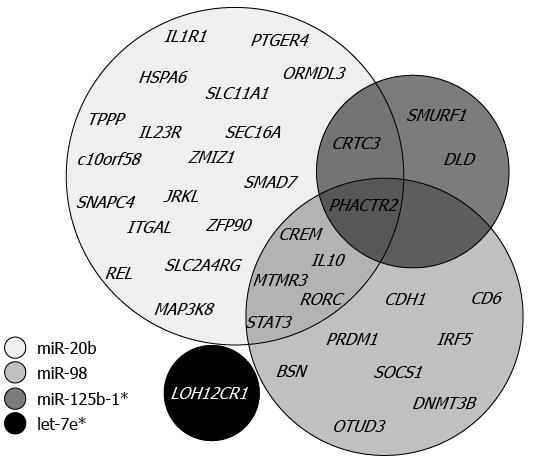
Figure 4 Venn diagram illustrating the miRNA-specific and overlapping ulcerative colitis associated target genes.
The four circles represent predicted miRNA targets found in the list of reported ulcerative colitis susceptibility genes.
- Citation: Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol 2013; 19(27): 4289-4299
- URL: https://www.wjgnet.com/1007-9327/full/v19/i27/4289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i27.4289