Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 7, 2007; 13(1): 91-103
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.91
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.91
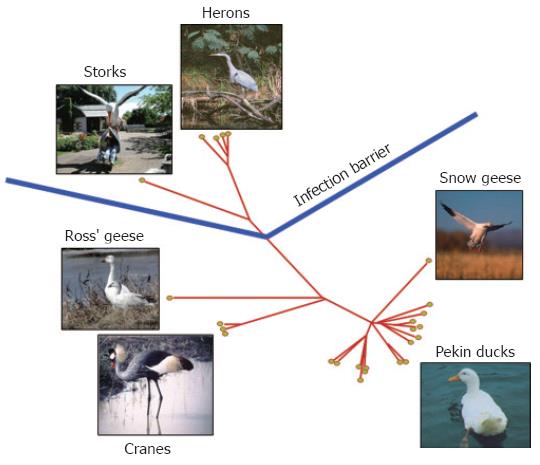
Figure 1 Phylogenetic tree of avian hepadnaviruses and related hosts.
Phylogenetic relationship of all known avian hepadnaviruses based on preS/S gene sequence. The corresponding natural hosts are also indicated.
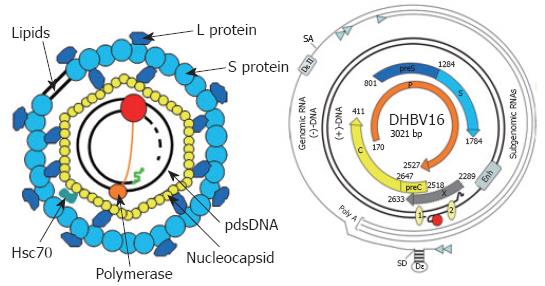
Figure 2 Virion structure and genome organization of avian hepadnaviruses.
The viral envelope is derived from hepatocellular membranes and contains the viral surface proteins S and L. It covers the nucleocapsid harbouring the viral genome with the covalently linked terminal protein domain (TP, orange circle) of the polymerase (P, red circle) and cellular proteins like Hsc70. The genome is organized as depicted. The various transcripts are indicated by thin lines with the small arrowhead indicating the start sites. The partially double stranded, viral DNA with the covalently bound TP domain of P (red circle) is symbolized by the thicker lines. The numbered circles 1 and 2 on the viral DNA represent the direct repeats (DR). Enh represents the enhancer domain. The ORFs encoding core (C), polymerase (P), and the surface proteins (preS and S) are symbolized by thick arrows. Epsilon (ξ) is the stem loop structure on the pgRNA which acts as an encapsidation signal and replication origin. The second encapsidation element DξII is unique to avian hepatitis B viruses, since the mammalian counterparts lack this RNA structure. SD and SA represent the major splice donor and acceptor sites, respectively.
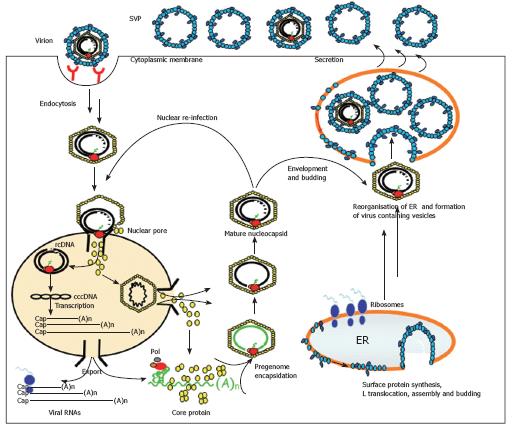
Figure 3 Model of the hepadnaviral life cycle.
The hepadnaviral particle binds the hepatocyte via a specific receptor complex/molecule. It then enters the cell via endocytosis and is transported intracellularly in a MT-dependent fashion. After release out of the endosomal compartment, the nucleocapsid containing the viral DNA binds the nuclear pore complex and the viral genome is released into the nucleus. It is also possible that the whole nucleocapsid enters the nucleus and releases the viral DNA inside. Afterwards, the viral rcDNA is converted into cccDNA and viral transcripts are made. These are exported into the cytosol where the pgRNA is packaged together with the viral polymerase into the nucleocapsid. Inside the capsid, the RNA is reverse transcribed into the viral DNA genome. These matured nucleocapsids can then either be transported to the nucleus to add to the cellular cccDNA pool or they interact with viral surface proteins on the cellular ER membrane. There, the nucleocapsid buds into the lumen of the ER and is then transported in a yet undefined fashion to the cell surface where the viral particle is released into the cell exterior.
Figure 4 Phosphorylation sites of the viral core protein.
The C-terminal sequence of the core proteins from DHBV and HBV are shown. Red brackets indicate the phosphorylation motifs, bold letters indicate the phosphorylated amino acid residue.
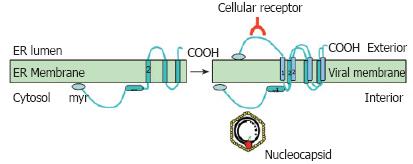
Figure 5 Dual topology of the viral surface protein L.
The L protein is inserted into the ER membrane during synthesis with transmembrane domain 2 being inserted into the membrane. Half of the proteins then change their topology and insert the transmembrane domain 1 into the ER membrane. After this change and formation of the virus, L can exert its two basic functions; interaction with the nucleocapsid on the cytosolic preS domain and interaction with the host cell receptor on the surface of the viral particle.
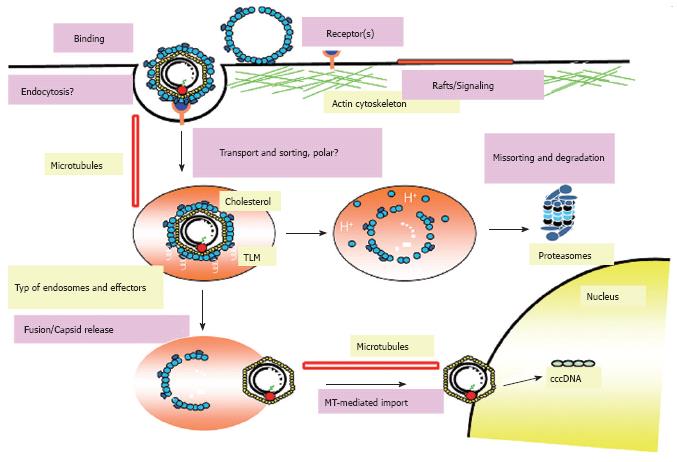
Figure 6 Current model for entry and intracellular trafficking of DHBV.
The hepadnaviral life cycle starts with the attachment of virions to specific binding sites on the surface of hepatocytes mediated by the preS region of the large viral envelope protein L. Afterwards the virus enters the cell via endocytosis and resides in an endocytic vesicle which is transported in a MT-dependent and probably polar fashion. The endosomal release of the incoming particle requires the activity of the vacuolar H+-ATPase and presumably the cholesterol of the viral envelope. After release, the particle is transported in a MT-dependent fashion mediated by dynein/dynactin to the nucleus and initiates infection after release of the viral genome at the nuclear membrane or within the nucleus. However, the majority of viral particles is missorted and degraded by the proteasome and other cellular proteases.
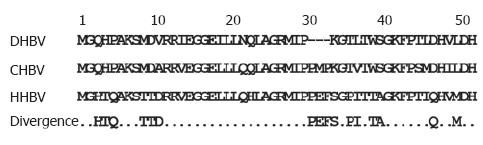
Figure 7 Sequence comparison of the viral L protein from different hepadnaviruses.
The first 50 amino acids of the preS domain of hepadnaviral L from duck, crane, and heron HBV are shown. The divergent amino acids are shown in the lowest lane.
- Citation: Funk A, Mhamdi M, Will H, Sirma H. Avian hepatitis B viruses: Molecular and cellular biology, phylogenesis, and host tropism. World J Gastroenterol 2007; 13(1): 91-103
- URL: https://www.wjgnet.com/1007-9327/full/v13/i1/91.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i1.91