Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Jan 7, 2007; 13(1): 14-21
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.14
Published online Jan 7, 2007. doi: 10.3748/wjg.v13.i1.14
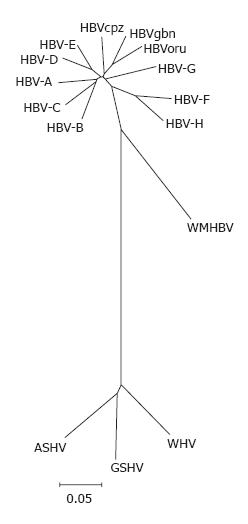
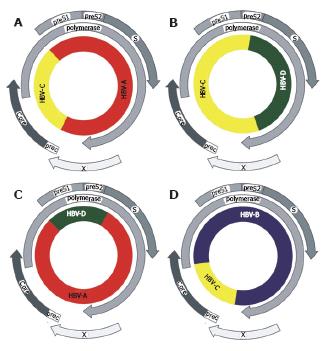
Figure 1 Phylogenetic tree of orthohe-padnaviruses.
Complete genomes of HBV genotypes A (X02763), B (D00330), C (M12906), D (V01460), E (X75657), F (X69798), G (AF160501) and H (AY090454); HBVcpz (D00220), HBVoru (NC 002168), and HBVgbn (U46935) were aligned using clustal w with orthohepadnavirus genomes from woolly monkey (AF046996) woodchuck (J02442), ground squirrel (K02715) and the tentative member from arctic squirrel (nc_001719). The alignment was tested with the neighbour-joining method.
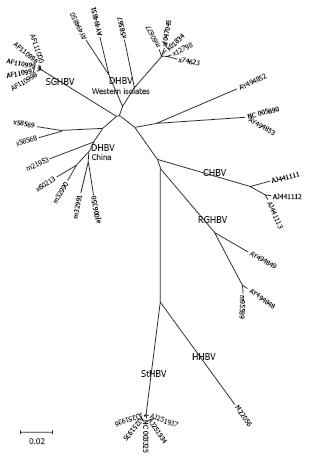
Figure 2 Phylogenetic tree of the genus avihepadnavirus.
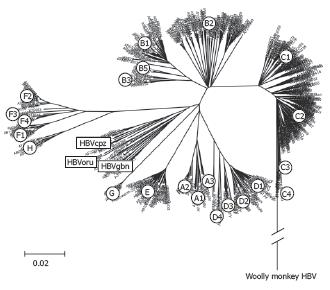
Figure 3 Phylogenetic tree of complete HBV genomes.
An alignment of 601 complete HBV sequences was performed with Clustal X in the program DNAstar. The alignment was further analysed by boot-strapping using the Neighbourhood-Joining method contained in MEGA version 3.1[104].
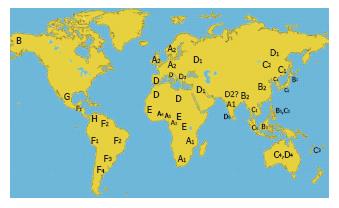
Figure 4 Geographic distribution of HBV genotypes and subgenotypes.
- Citation: Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol 2007; 13(1): 14-21
- URL: https://www.wjgnet.com/1007-9327/full/v13/i1/14.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i1.14