Copyright
©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Jun 7, 2006; 12(21): 3344-3351
Published online Jun 7, 2006. doi: 10.3748/wjg.v12.i21.3344
Published online Jun 7, 2006. doi: 10.3748/wjg.v12.i21.3344
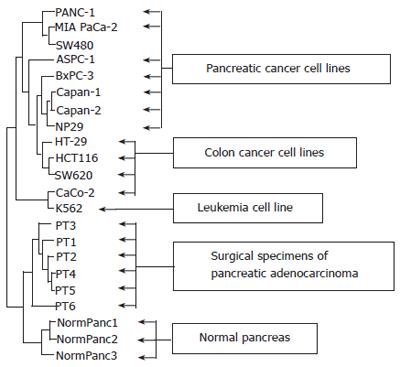
Figure 1 Hierarchical clustering analysis of sample expression profiles.
Dendrogram of centered mean expression data using euclidian distance with average linkage clustering, showing relationships in gene expression profiles of samples. Closely related samples are found in the same branch of the tree and a reduced branch height represents a closer relationship between groups.
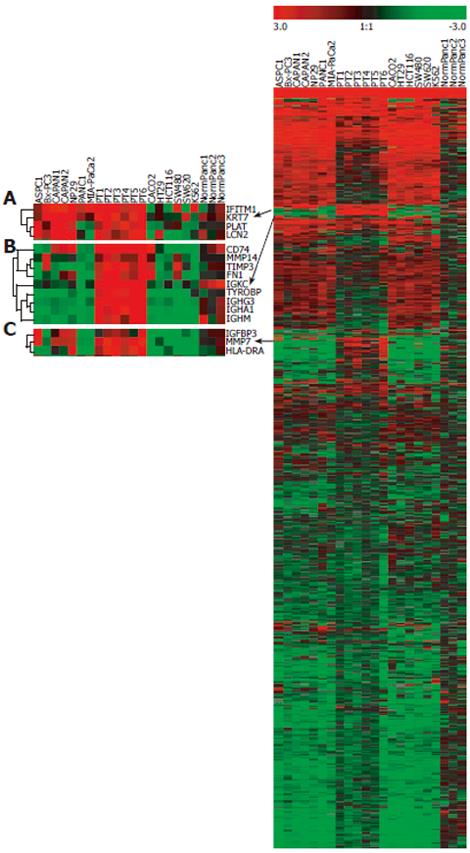
Figure 2 Hierarchical clustering analysis of gene expression profiles.
Right: Expression profile of the 871 genes retained after data-filtering. Left: Three magnified subclusters (A-C) showing the genes overexpressed in most of pancreatic tumors. Red color indicates high expression levels; green color indicates low expression levels.
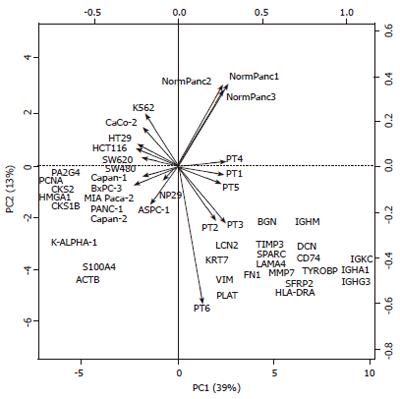
Figure 3 Principal component analysis (PCA) of gene expression data.
Biplot resulting from a PCA of the line and column centered data containing 871 genes (individuals) in lines and 22 samples in columns (variables). The 28 genes contributing the most to the total variability are shown. The two principal components (PC1 and PC2) contribute to more than 50% (39% and 13%) of the total variability and resolve four biological sample categories: PC1 on the horizontal x-axis distinguish between cell lines (left) and tissue samples (right) whereas PC2 on the vertical y-axis distinguish between malignant pancreas samples (bottom) and other sample categories (top).
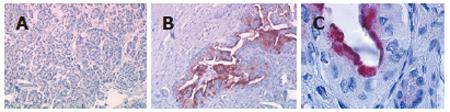
Figure 4 Lipocalin 2 protein expression in pancreatic cancer tissue Immunohistochemical analysis by AEC + substrate chromogen staining.
A (x 40): normal pancreas; B (x 200), C (x 1000): pancreatic adenocarcinomas.
- Citation: Laurell H, Bouisson M, Berthelémy P, Rochaix P, Déjean S, Besse P, Susini C, Pradayrol L, Vaysse N, Buscail L. Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J Gastroenterol 2006; 12(21): 3344-3351
- URL: https://www.wjgnet.com/1007-9327/full/v12/i21/3344.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i21.3344