Copyright
©The Author(s) 2004.
World J Gastroenterol. Apr 15, 2004; 10(8): 1082-1087
Published online Apr 15, 2004. doi: 10.3748/wjg.v10.i8.1082
Published online Apr 15, 2004. doi: 10.3748/wjg.v10.i8.1082
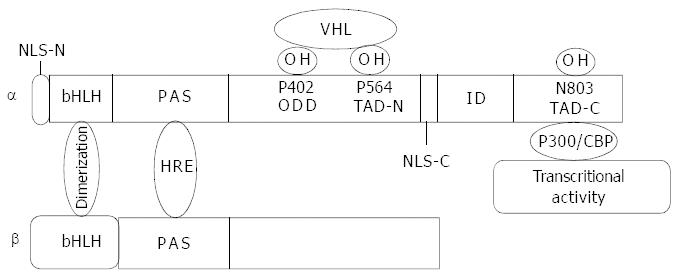
Figure 1 Molecular structure of HIF-1α and HIF-1β.
bHLH domain mediates dimerization of the two subunits. PAS domain is responsible for DNA binding. Proline residues of 402 and 564 at ODD domain are hydroxylized by proline hydroxylase and recog-nized by VHL and then targeted to the ubiquitin proteasome pathway. Asn803 at the C-terminal transactivation domain (TAD-C) is hydroxylized by FIH-1 (factor inhibiting HIF-1) with a result of inhibition of HIF-1α interaction with co-activator p300 and conse-quently inhibits transcriptional activity. The nuclear location signal at C-terminal functions in HIF-1α translocation into nuclei.
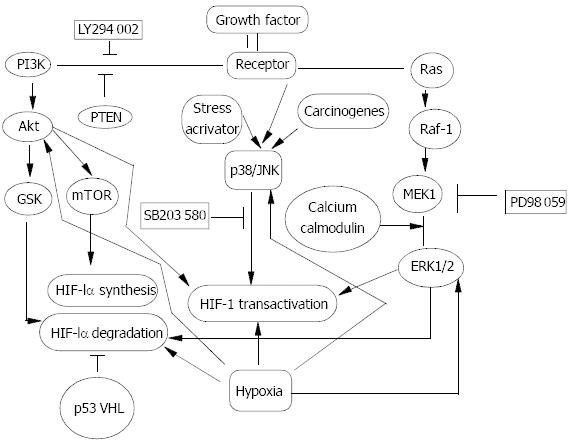
Figure 2 Signal transduction pathway in HIF-1α regulation.
Oncogenes, growth factors and hypoxia have been documented to regulate HIF-1α protein and increase its transactivity. PI-3K/Akt, Raf-1/MEK/ERK1/2 and p38/JNK pathways were activated in response to oncogenes, growth factors and stress activators such as hypoxia, and carcinogens to different ex-tent in a cell type- and stimulus type-specific manner. GSK and mTOR were two target events of Akt and could contribute to decreasing HIF-1α degradation and increasing HIF-1α pro-tein synthesis. Activated ERK1/2 could mainly up-regulate
- Citation: Shi YH, Fang WG. Hypoxia-inducible factor-1 in tumour angiogenesis. World J Gastroenterol 2004; 10(8): 1082-1087
- URL: https://www.wjgnet.com/1007-9327/full/v10/i8/1082.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i8.1082