Copyright
©The Author(s) 2015.
World J Radiol. May 28, 2015; 7(5): 104-109
Published online May 28, 2015. doi: 10.4329/wjr.v7.i5.104
Published online May 28, 2015. doi: 10.4329/wjr.v7.i5.104
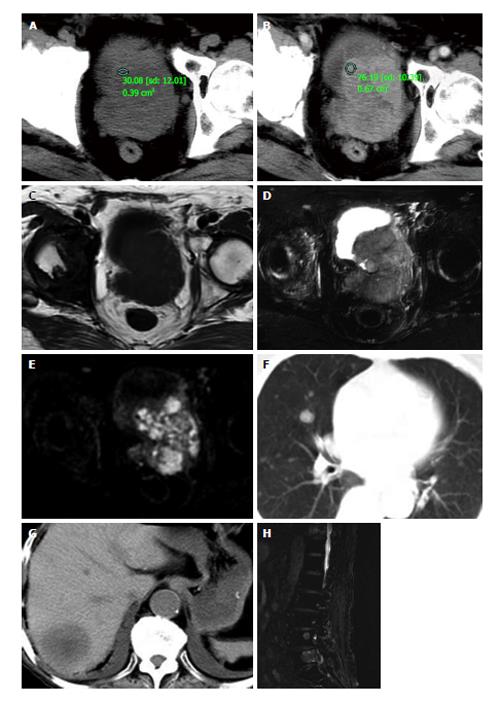
Figure 1 The diagnosis.
Precontrast (A) and postcontrast (B) computed tomography (CT) images show the prostate approximately 6.5 cm × 7.5 cm × 7.2 cm in size with an irregular border and small necrosis in the center region, the solid part enhanced obviously in arterial phase of contrast enhancement CT scanning. T1-weighted image (T1WI, C) show the lesion to be slightly heterogeneous low signal intensity, T2WI (D), and diffusion-weighted image (E) show the same region to be heterogeneous hyperintense. Chest (F), abdominal (G) CT and lumbar (H) magnetic resonance imaging revealed multiple metastases to lungs, liver, and lumbar vertebraes.
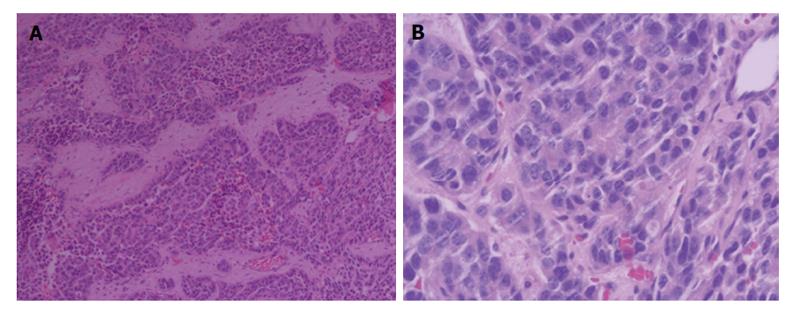
Figure 2 Microscopic finding showed infiltrating nests of small cells in fibrotic stroma.
Tumor cells had small hyperchromatic nuclei and scanty cytoplasms. H and E: × 100 (A), and × 200 (B).
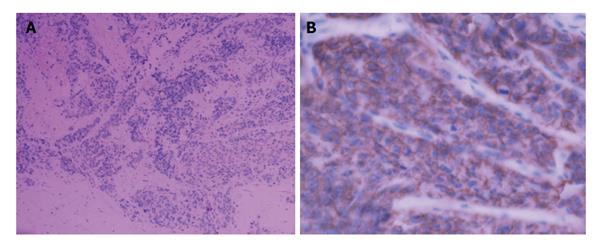
Figure 3 Immunohistochemical stain showed positivity of tumor cells for CgA (A) and a positive staining with CD56, CD56 (+) (B).
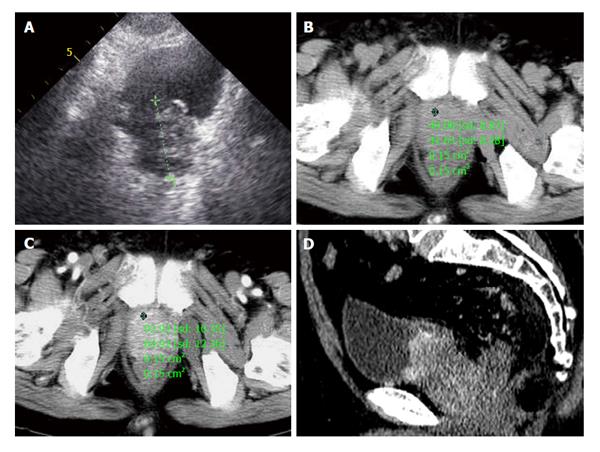
Figure 4 A 72-year-old male patient presented with one month of dysuria.
A: Prostate enlarged to 4.0 cm × 5.1 cm × 4.2 cm with incomplete envelope, irregular shape and heterogeneous echo texture; B: A pre-enhanced computed tomography (CT) scan image of the pelvis showed a marked enlarged prostatic tumor invading to bladder, seminal vesicle; C: The enlarged prostate was inhomogeneous moderately enhancement in arterial phase; D: Coronal oblique multiplanar reconstruction of enhanced CT scan shows the enhanced inhomogeneous prostate with high attenuation protruding into bladder and unclear with surrounding tissue.
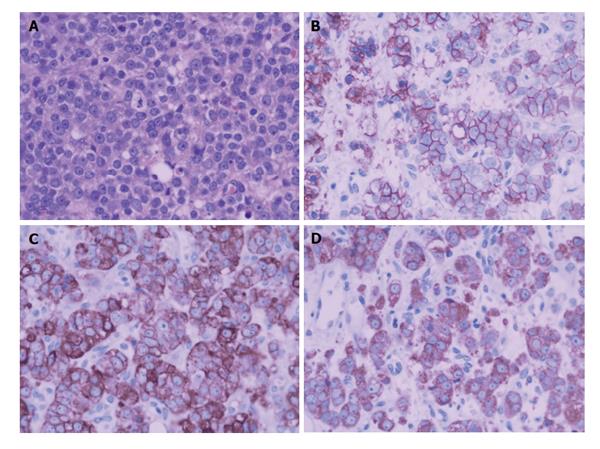
Figure 5 A biopsy of the prostate mass.
Microscopic findings (A) showed infiltrating nests of small cells in fibrotic stroma. Tumor cells had small hyperchromatic nuclei and scanty cytoplasms (H and E, × 200). Immunohistochemical stain showed strong positivity of tumor cells for CD56(++) (B), CgA(++) (C), Syn(++) (D).
- Citation: He HQ, Fan SF, Xu Q, Chen ZJ, Li Z. Diagnosis of prostatic neuroendocrine carcinoma: Two cases report and literature review. World J Radiol 2015; 7(5): 104-109
- URL: https://www.wjgnet.com/1949-8470/full/v7/i5/104.htm
- DOI: https://dx.doi.org/10.4329/wjr.v7.i5.104