Copyright
©2013 Baishideng Publishing Group Co.
World J Cardiol. Aug 26, 2013; 5(8): 305-312
Published online Aug 26, 2013. doi: 10.4330/wjc.v5.i8.305
Published online Aug 26, 2013. doi: 10.4330/wjc.v5.i8.305
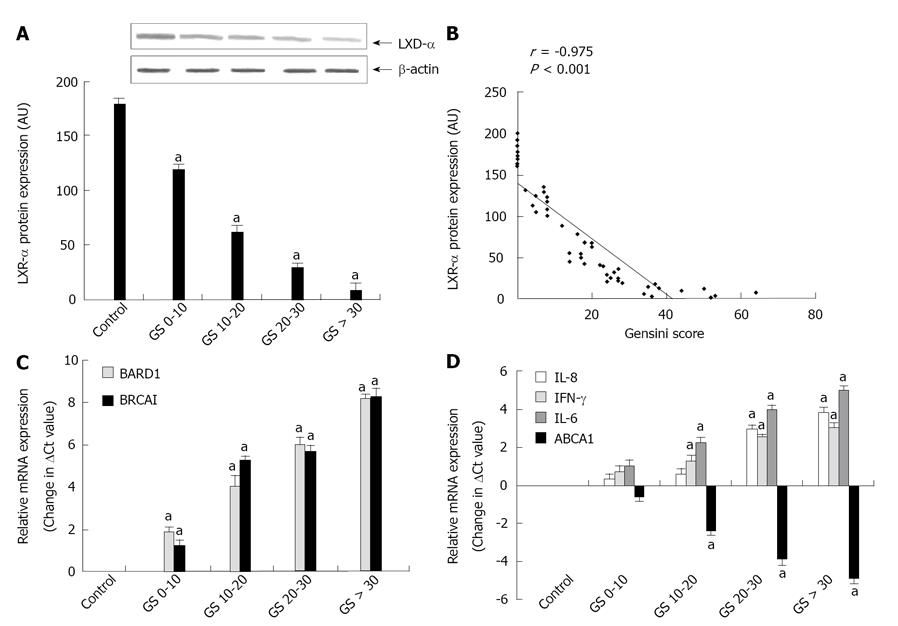
Figure 1 Gene expression analysis of various genes with respect to gensini score.
A: Mean values of liver X receptor-α (LXR-α) protein levels in peripheral blood mononuclear cells (PBMCs) isolated from coronary heart disease (CHD) subjects and healthy controls with respect to increasing gensini score (reflecting the severity of coronary occlusion). Each bar represents mean ± SD for 10 different individuals in each group. The means were compared with one way ANOVA and aP < 0.05 vs control group; B: Statistical correlation between translational expression of mutant LXR-α and gensini score within PBMCs. Values of r show Spearman rank correlation coefficient and P < 0.01 was considered statistically significant; C, D: Mean values of [breast and ovarian cancer susceptibility1 (BRCA1)-associated RING domain 1] (BARD1) and BRCA1 (C) and interleukin (IL)-8, interferon (IFN)-γ, IL-6, and ATP-binding cassette A1 (ABCA1) (D) mRNA expression (change in ΔCt values) in PBMCs derived from CHD patients and healthy controls with respect to increasing Gensini Score. Each bar represents mean ± SD for 10 different individuals in each group. The means were compared with one way ANOVA and aP < 0.05 vs control group.
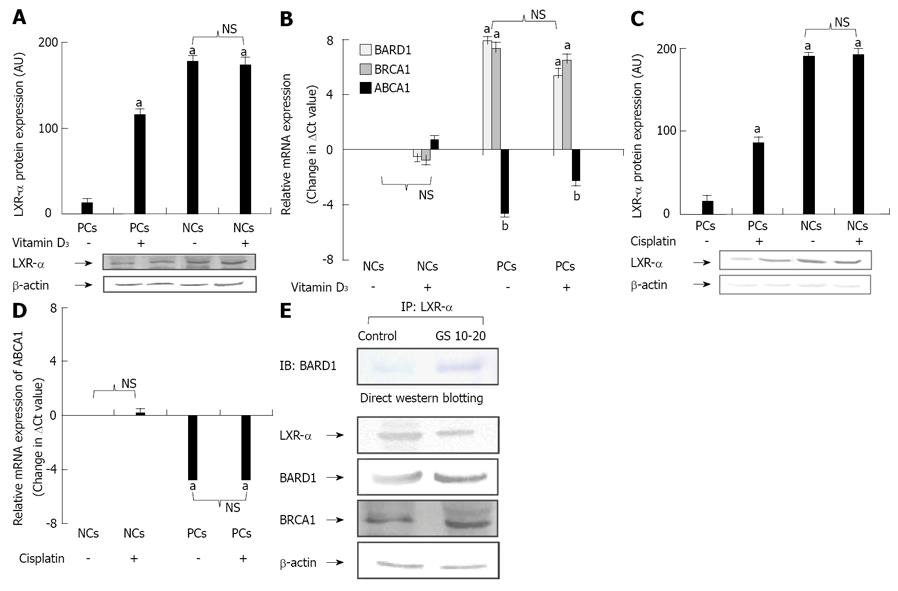
Figure 2 Expression analysis of various genes upon exposure to Vitamin D3 and Cisplatin in peripheral blood mononuclear cells of coronary heart diseasesubjects and their healthy counterparts.
A: Protein expression of liver X receptor-α (LXR-α) within peripheral blood mononuclear cells (PBMCs), isolated from coronary heart disease (CHD) subjects (GS > 30) as well as normal healthy controls, exposed to culture medium enriched with and without Vitamin D3 (1 μmol/L); B: Relative mRNA expression (change in ΔCt values) of [breast and ovarian cancer susceptibility 1 (BRCA1)-associated RING domain 1] (BARD1), BRCA1 and ATP-binding cassette A1 (ABCA1) upon Vitamin D3 exposure in normal and patient cells; C: Protein expression of LXR-α within PBMCs, isolated from CHD subjects (GS > 30) as well as normal healthy controls, exposed to culture medium enriched with and without Cisplatin (30 μmol/L); D: Relative mRNA expression (change in ΔCt values) of ABCA1 upon Cisplatin exposure in normal and patient cells. Each bar represents mean ± SD for the combined results of three independent experiments from different individuals in triplicate. The means were compared with one way ANOVA and aP < 0.05 vs control group (A-D); E: Total cell lysates from PBMCs derived from CHD subjects and normal healthy individuals were immunoprecipitated with anti LXR-α antibody and immunoblotted with anti-BARD1 antibody. The direct western blotting shows the expression of LXR-α, BARD1, BRCA1 and β-actin (Sigma Aldrich) in PBMCs of CHD subjects and normal healthy controls. The experiments were repeated three times from different individuals and representative results are shown. IB: Immunoblotting; IP: Immunoprecipitation; NC: Normal cells; NS: Non-significant; PC: Patient cells.
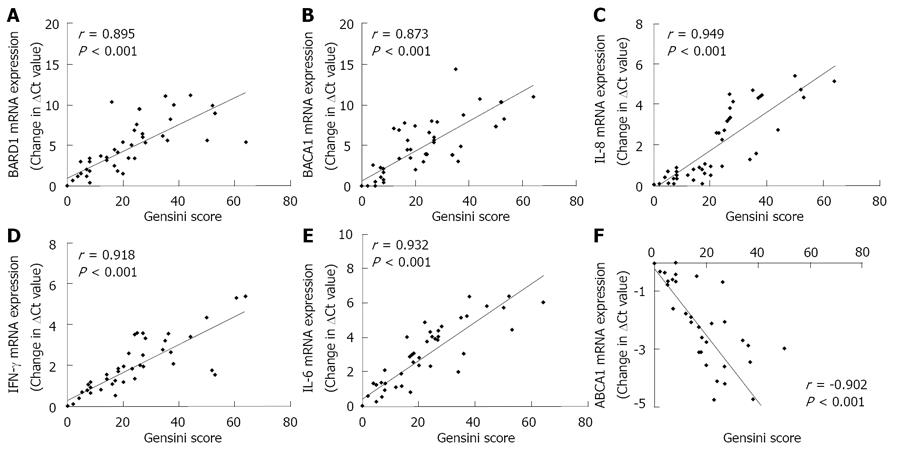
Figure 3 Correlation of mRNA expression of various genes with the gensini score.
Statistical correlation between transcriptional expression of [breast and ovarian cancer susceptibility1 (BRCA1)-associated RING domain1] (BARD1) (A), BRCA1 (B), interleukin (IL)-8 (C), interferon (IFN)-γ (D), IL-6 (E) and ATP-binding cassette A1 (ABCA1) (F) and gensini score within peripheral blood mononuclear cells derived from CHD subjects and normal healthy controls. Values of r show Spearman rank correlation coefficient.
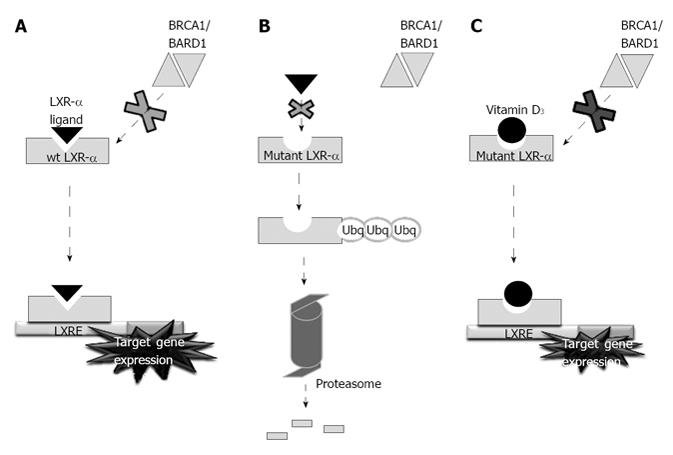
Figure 4 Schematic diagram representing the mechanism of action of liver X receptor-α.
A: Wild type functional liver X receptor-α (LXR-α) in healthy controls; B: Mutant non-functional LXR-α in coronary heart disease patients; C: Mutated but functional (to some extent) LXR-α when rescued by Vitamin D3. BRCA1: Breast and ovarian cancer susceptibility 1; BARD1: BRCA1-associated RING domain 1; LXRE: liver X receptor.
- Citation: Arora M, Kaul D, Sharma YP. Blood cellular mutant LXR-α protein stability governs initiation of coronary heart disease. World J Cardiol 2013; 5(8): 305-312
- URL: https://www.wjgnet.com/1949-8462/full/v5/i8/305.htm
- DOI: https://dx.doi.org/10.4330/wjc.v5.i8.305