Copyright
©2012 Baishideng Publishing Group Co.
World J Biol Chem. Mar 26, 2012; 3(3): 53-60
Published online Mar 26, 2012. doi: 10.4331/wjbc.v3.i3.53
Published online Mar 26, 2012. doi: 10.4331/wjbc.v3.i3.53
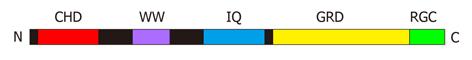
Figure 1 A schematic representation of human IQGAP1.
The domains are shown on a linear representation of the protein sequence. The folded protein is unlikely to be arranged in a linear fashion and it is possible that some of these domains contact each other. CHD: Calponin homology domain (residues 44-159); WW: WW domain (679-712); IQ: IQ-motifs containing region (745-864); GRD: GAP-related domain (1004-1237); RGC: Ras-GAP C-terminal domain (1563-1657). The definitions of the domain boundaries are those of Briggs and Sacks[2].
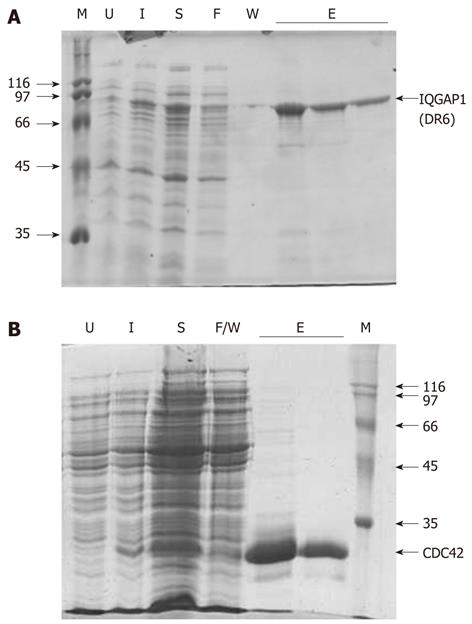
Figure 2 Proteins used in this study.
The recombinant expression and purification of (A) IQGAP1(DR6) and (B) CDC42 monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In both (A) and (B), U, extract from cells prior to induction; I, extract from cells 2-3 h after induction; S, soluble material remaining after sonication; F, material which flowed through the column without binding; W, material removed in the washing steps; E, elutions; M, molecular mass markers (with their masses shown to the side of the gel in kDa). In the case of CDC42, F and W were combined into a single sample.
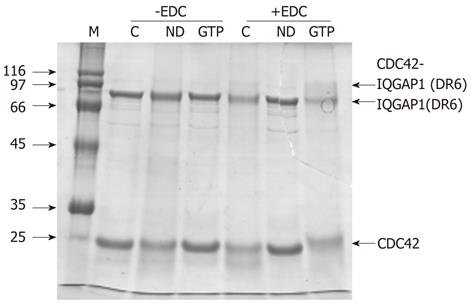
Figure 3 Interaction between IQGAP1 and CDC42.
An interaction between IQGAP1(DR6) and CDC42 can be detected by EDC crosslinking in the presence of GTP. In the absence of EDC (lanes headed -EDC), a mixture of IQGAP(DR6) and CDC42 (lane C) behaves the same on SDS-PAGE as IQGAP1(DR6) and nucleotide-depleted CDC42 (lane ND) and as IQGAP1(DR6) and GTP-loaded CDC42 (lane GTP). In the presence of the crosslinker (lanes headed +EDC), crosslinking was not observed between IQGAP(DR6) and CDC42 (C) or IQGAP(DR6) and nucleotide-depleted CDC42 (ND). However an additional band, corresponding to approximately the combined molecular masses of IQGAP(DR6) and CDC42 is seen with GTP-loaded CDC42 (GTP).
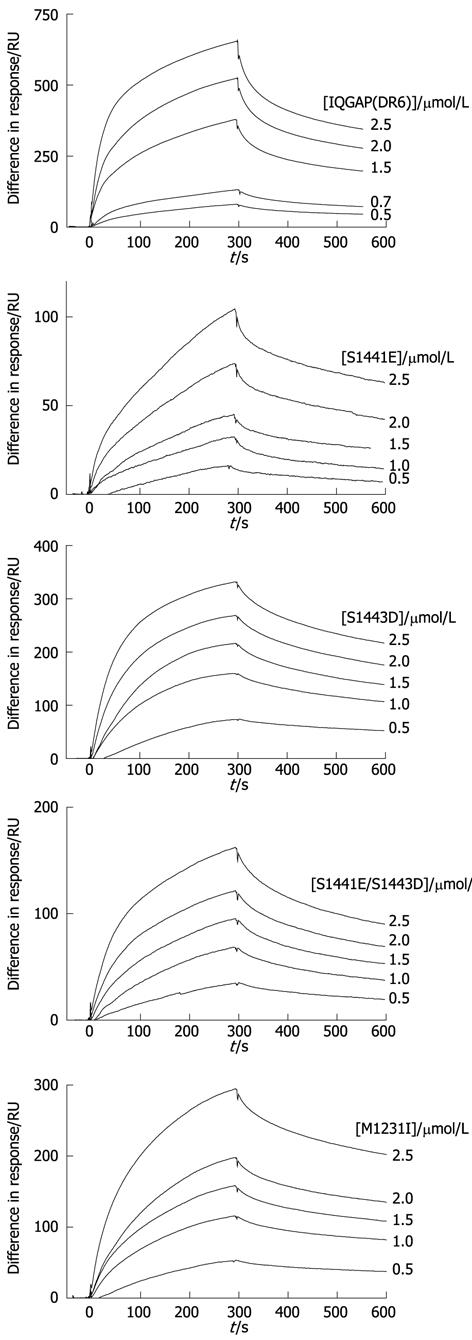
Figure 4 Interaction of IQGAP-CTD and CDC42 can be detected by surface plasmon resonance.
Typical sensorgrams resulting from flowing IQGAP1(DR6) wild type and variants over immobilised CDC42 (for conditions, see Materials and Methods). From top to bottom, wild type, S1441E, S1443D, S1441E/S1443D, M1231I. Protein concentrations are shown to the right of the sensorgrams.
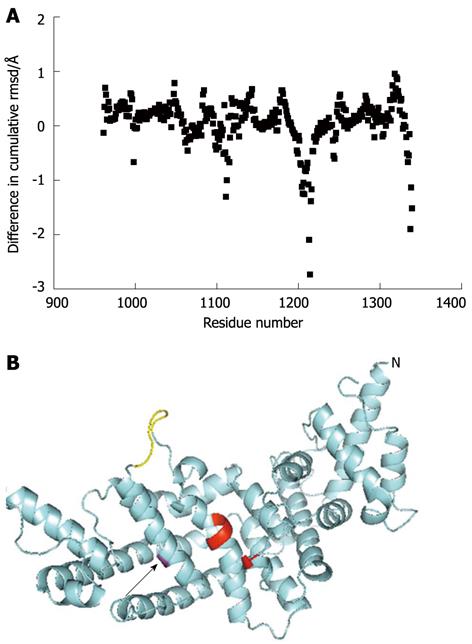
Figure 5 Predicted structural consequences of the M1231I, cancer associated, mutation.
A: The predicted effects on backbone flexibility as determined by FIRST/FRODAN (see Materials and Methods) are plotted as the difference in cumulative flexibility between the M1231I variant and wild type. Thus, a negative value represents in loss in flexibility of the variant compared to wild type. The greatest predicted rigidification occurs around residue 1214 and is indicated with an arrow; B: A model of the GAP-related domain of the M1231I variant is shown in cyan, with Ile-1231 in magenta and indicated with an arrow. The N-terminus of the fragment is marked (N) and the C-terminus can be seen close to this in space. Key residues predicted to be involved in CDC42 interaction (Thr-1046 and Tyr-1192 to Arg-1194) are shown in red. A loop (Ser-1212 to Leu-1217) predicted to be more rigid in the variant compared to the wild type is shown in yellow. It links the helix containing residue 1231 and part of the CDC42 binding site.
- Citation: Elliott SF, Allen G, Timson DJ. Biochemical analysis of the interactions of IQGAP1 C-terminal domain with CDC42. World J Biol Chem 2012; 3(3): 53-60
- URL: https://www.wjgnet.com/1949-8454/full/v3/i3/53.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v3.i3.53