Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1437
Peer-review started: October 13, 2023
First decision: December 8, 2023
Revised: December 19, 2023
Accepted: January 17, 2024
Article in press: January 17, 2024
Published online: April 15, 2024
Gastric cancer, characterized by a multifactorial etiology and high heterogeneity, continues to confound researchers in terms of its pathogenesis. Curcumin, a natural anticancer agent, exhibits therapeutic promise in gastric cancer. Its effects include promoting cell apoptosis, curtailing tumor angiogenesis, and enhancing sensitivity to radiation and chemotherapy. Long noncoding RNAs (lncRNAs) have garnered significant attention as biomarkers for early screening, diagnosis, treatment, and drug response because of their remarkable specificity and sensitivity. Recent investigations have revealed an association between aberrant lncRNA expression and early diagnosis, clinical staging, metastasis, drug sensitivity, and prognosis in gastric cancer. A profound understanding of the intricate mechanisms through which lncRNAs influence gastric cancer develop
To identify lncRNAs associated with curcumin treatment and investigate the role of lncRNA AC022424.2 in the effects of curcumin on gastric cancer cell apoptosis, proliferation, and invasion. Furthermore, these findings were validated in clinical samples.
The study employed CCK-8 assays to assess the impact of curcumin on gastric cancer cell proliferation, flow cytometry to investigate its effects on apoptosis, and scratch and Transwell assays to evaluate its influence on the migration and invasion of BGC-823 and MGC-803 cells. Western blotting was used to gauge changes in the protein expression levels of CDK6, CDK4, Bax, Bcl-2, caspase-3, P65, and the PI3K/Akt/mTOR pathway in gastric cancer cell lines after curcumin treatment. Differential expression of lncRNAs before and after curcumin treatment was assessed using lncRNA sequencing and validated using quantitative reverse transcription polymerase chain reaction (qRT-PCR) in BGC-823 and MGC-803 cells. AC022424.2-1 knockdown BGC-823 and MGC-803 cells were generated to scrutinize the impact of lncRNA AC022424.2 on apoptosis, proliferation, migration, and invasion of gastric cancer cells. Western blotting was performed to ascertain changes in the expression of proteins implicated in the PI3K/Akt/mTOR and NF-κB signaling pathways. RT-PCR was employed to measure lncRNA AC022424.2 expression in clinical gastric cancer tissues and to correlate its expression with clinical pathological characteristics.
Curcumin induced apoptosis and hindered proliferation, migration, and invasion of gastric cancer cells in a dose- and time-dependent manner. LncRNA AC022424.2 was upregulated after curcumin treatment, and its knockdown enhanced cancer cell aggressiveness. LncRNA AC022424.2 may have affected cancer cells via the PI3K/Akt/mTOR and NF-κB signaling pathways. LncRNA AC022424.2 downregulation was correlated with lymph node metastasis, making it a potential diagnostic and prognostic marker.
Curcumin has potential anticancer effects on gastric cancer cells by regulating lncRNA AC022424.2. This lncRNA plays a significant role in cancer cell behavior and may have clinical implications in diagnosis and prognosis evaluation. The results of this study enhance our understanding of gastric cancer development and precision treatment.
Core Tip: We investigated the effects of curcumin on gastric cancer cells, and we found that curcumin inhibited gastric cancer cell growth and induced apoptosis via long noncoding RNA (lncRNA) AC022424.2. Furthermore, the AKT and mTOR signaling pathways were involved in the process. We validated the role of lncRNA AC022424.2 on clinical samples and found that lncRNA AC022424.2 is associated with lymph metastasis.
- Citation: Wang BS, Zhang CL, Cui X, Li Q, Yang L, He ZY, Yang Z, Zeng MM, Cao N. Curcumin inhibits the growth and invasion of gastric cancer by regulating long noncoding RNA AC022424.2. World J Gastrointest Oncol 2024; 16(4): 1437-1452
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1437.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1437
Gastric cancer (GC) is a highly heterogeneous tumor with a global overall survival rate of approximately 25%[1]. Most GC cases are diagnosed at advanced stages, with approximately 60% of patients presenting with local or distant metastasis at the time of diagnosis[2]. Diagnosis involves qualitative and locational determination through endoscopic biopsies, and staging is performed using various techniques, including computed tomography, endoscopic ultrasonography, positron emission tomography, and laparoscopy. The clinical staging of GC determines whether the treatment approach is curative or palliative. Deep learning techniques for artificial intelligence analysis of whole-slide histopathology images can rapidly detect tumors in gastric biopsies and resection specimens, offering high sensitivity and specificity[3-6]. Liquid biopsy techniques include the detection of cell-free circulating tumor DNA (ctDNA), circulating tumor cells, exosomes, and tumor tissue-derived platelets. This technology holds promise for early GC diagnosis, disease monitoring, drug response prediction, and disease recurrence identification. Moreover, compared with tissue biopsies, liquid biopsies provide a more comprehensive view of the high heterogeneity of GC[7].
Tumor metastasis, the high heterogeneity of cancer cells, and chemotherapy resistance are primary factors that contribute to the poor overall prognosis of GC[8]. Despite improvements in public health conditions and effective eradication of Helicobacter pylori in some regions, GC remains a fatal disease. Current treatment approaches for GC include surgery, traditional chemotherapy, targeted therapy, and immunotherapy; however, these strategies have limited effectiveness in controlling the disease. Chemotherapy is a key strategy for treating mid- to late-stage GC[9]; however, its role in GC treatment is constrained by adverse drug reactions and drug resistance. Chinese herbal medicine, which is rooted in a tradition spanning millennia, has been a cornerstone of healthcare in China and across Asia. A distinctive feature of Chinese herbal medicine is the use of herbal formulas, which comprise multiple herbs, to improve abnormal symptoms associated with specific diseases[10]. In the realm of exploring alternative cancer treatments, natural compounds with anticancer properties, such as curcumin, are garnering attention because of their low adverse effects. Although the anticancer effects of curcumin have been extensively studied, its specific mechanisms of action remain incompletely understood.
Curcumin is a biologically active compound with antibacterial properties. In 1985, Kuttan et al[11] confirmed its anticancer effects. Curcumin exerts anticancer effects by inhibiting cell cycle progression and promoting apoptosis[12]. Despite substantial research confirming the anticancer effects of curcumin, its precise mechanisms of action remain partially elucidated. Gene mutations play a critical role in GC occurrence and development. Mutations do not solely occur in the coding regions of the cancer cell genome; they predominantly occur in vast noncoding regions. With significant advances in DNA sequencing technology, research on the noncoding regions of cancer cells has become a research hotspot. Recently, several long noncoding RNAs (lncRNAs) with high expression have been discovered in various cancers, including GC, colorectal cancer, liver cancer, and lung cancer. LncRNAs participate in multiple cancer signaling pathways, such as Notch, mTOR, NF-κB, and Wnt[13-15]. They regulate cell proliferation, migration, apoptosis, invasion, cell cycle, and metastasis. Numerous lncRNAs longer than 200 base pairs have been identified as crucial components of cancer biology. Recently, the differential expression of certain lncRNAs in GC has been progressively discovered.
This study employs lncRNA sequencing to screen for differentially expressed lncRNAs after curcumin treatment. Further investigation will explore the relationship between the ability of curcumin to inhibit GC cells and lncRNA AC022424.2. This study reveals how curcumin regulates the apoptosis, proliferation, and invasion of GC cells through lncRNA modulation. The subsequent validation of these experimental results in clinical tissue specimens and exploration of specific mechanisms will provide more theoretical support for elucidating the role of lncRNAs in the occurrence and development of GC and identifying precise therapeutic targets.
The BGC823 and MGC803 human gastric adenocarcinoma cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). BGC823 cells were cultured in RPMI-164 and MGC803 cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco, Thermo Fisher Scientific, Waltham, MA, United States), supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mmol/L L-glutamine, and 100 U/mL penicillin/streptomycin (HyClone, Logan, UT, United States), at 37°C with 5% CO2 in a humidified incubator.
The curcumin compound (Solarbio, Beijing, China) was dissolved in dimethyl sulfoxide (DMSO) to make the stock solution (5 mmol/L), and various concentrations were prepared by diluting in culture medium.
Fifty-one GC tissues and matched adjacent normal gastric mucosa tissues were collected from GC patients who underwent gastrectomy from March 2018 to March 2019 at the First Affiliated Hospital, Lanzhou University. All cancer tissues were confirmed to be gastric adenocarcinomas by a pathologist using hematoxylin and eosin slides. Patients receiving radiotherapy, chemotherapy, or immunotherapy were excluded. Written informed consent was obtained from each patient, and the protocols for human sample handling were approved by the Ethics Review Committee of Lanzhou University.
BGC823 cells were treated with curcumin (40 μmol/L) for 24 h; as a negative control, the cells were treated with DMSO. Total RNA was extracted for lncRNA sequencing to identify differentially expressed lncRNAs.
For knocking down lncRNA AC022424.2, three plasmids of shRNA targeting different sequences of lncRNA AC022424.2-40921: TGTCCTTGCCTGTGGTGTCAA, AC022424.2-40922: TACCTTCATGAGAACCACAA, and AC022424.2-40923: TACCTTCATGAGAACCACAA were constructed by inserting these oligonucleotides into the ORF of lentiviral Lv-006 plasmid via the EcoRI and AgeI restriction sites. BGC823 cells were seeded at 2 × 104 cells per well in 6-well plates and cultured overnight. The cells were then transfected with the above plasmids using Lipofectamine 3000 (Invitrogen, Waltham, MA, United States) according to the manufacturer’s instructions.
Total RNA was extracted from cells and homogenized tissues using the MolPure® Cell/Tissue Total RNA Kit (Yeasen Biotechnology, Shanghai, China). cDNA was synthesized using the Hifair® II 1st Strand cDNA Synthesis Kit (gDNA digester plus; Yeasen Biotechnology). Real-time PCR was conducted using Hieff® qPCR SYBR® Green Master Mix (Yeasen Biotechnology) with the primers listed in Table 1 according to the manufacturer’s instructions.
| Clinical pathological features | Number of cases | AC022424.2 expression, mean ± SEM | P value |
| Sex | 0.740 | ||
| Male | 37 | 0.068 ± 0.032 | |
| Female | 14 | 0.050 ± 0.020 | |
| Age in yr | 0.856 | ||
| ≥ 60 | 32 | 0.067 ± 0.037 | |
| < 60 | 19 | 0.058 ± 0.024 | |
| Site | 0.508 | ||
| Esophagus | 11 | 0.030 ± 0.013 | |
| Gastric body/antrum | 40 | 0.071 ± 0.030 | |
| Differentiation grade | 0.617 | ||
| High–moderate differentiation | 3 | 0.021 ± 0.016 | |
| Low–poor differentiation | 48 | 0.067 ± 0.026 | |
| Clinical stage | 0.285 | ||
| I–II | 18 | 0.098 ± 0.062 | |
| III–IV | 33 | 0.044 ± 0.015 | |
| Lymph node metastasis | 0.021 | ||
| Yes | 35 | 0.014± 0.003 | |
| No | 16 | 0.032± 0.008 | |
| T stage | 0.311 | ||
| T1, T2 | 12 | 0.014 ± 0.004 | |
| T3, T4 | 39 | 0.076 ± 0.030 |
Cells were seeded into 96-well plates with 2000 cells/well in 100 μL medium and cultured at 37 °С. At the indicated time points, cell viability was detected using the CCK-8 kit (Yeasen Biotechnology). Specifically, 10 μL of CCK-8 solution was added to each well, followed by incubation for 2 h at 37 °С. Absorbance was then measured at 450 nm using a Microplate Reader Model-1680 (Bio-Rad, Hercules, CA, United States). Six replicates were set for each treatment.
Cells were seeded in 6-well plates and cultured until 70% confluence, followed by the addition of curcumin (40 μmol/L) or DMSO and incubation for an additional 6 h. The cells were then collected and stained with fluorescein isothiocyanate-conjugated anti-annexin V and propidium iodide (Yeasen Biotechnology) for 15 min, followed by washing with phosphate-buffered saline. Flow cytometric analysis was then performed. Each sample was tested in triplicate.
Cells were seeded at 5 × 104 cells/well into 6-well plates and cultured in standard medium with 1% FBS overnight. Wounds were created by scratching the cell monolayer using a sterile pipette tip, and the images were taken for the 0 h time point. The cells were cultured, and images were taken at 6, 12, 24, and 36 h. Three replicates were imaged for each plate.
Cells were seeded at 3 × 105 cells/well into the upper wells of Transwell plates, and 600-μL culturing medium with 15% FBS was added into the lower wells. The system was then cultured in a cell incubator for 24 h. The filters were fixed with 4% paraformaldehyde. The cells on the upper side of the filters were wiped with cotton swabs, and cells that migrated to the lower side of the filters were stained with crystal violet and photographed for analysis. For the Transwell invasion assay, cells were seeded at 20000 cells/well into the upper wells of 24-well plates that were coated with Matrigel, and culture medium with 20% FBS was added to the lower wells. Cells that invaded through the Matrigel were photographed after 48 h.
BGC-823 and MGC-803 cells were seeded in 6-well plates at 3 × 105 cells/well. After 24 h, the cells were treated with curcumin (40 μmol/L) for 6 h and lysed with RIPA buffer (Solarbio, China). Protein concentration was measured using the BCA Protein Assay Kit (Solarbio, China) according to the manufacturer’s instructions. Proteins were separated using SDS-PAGE and transferred onto polyvinylidene fluoride membranes (GE Healthcare, Chicago, IL, United States). The blots were incubated with the primary antibodies overnight at 4 °С and subsequently with secondary antibodies at room temperature for 1 h. The proteins were visualized using Super ECL Detection Reagent (Yeasen Biotechnology). An antibody against GAPDH was used as the loading control.
All statistical analyses were performed using GraphPad Prism (version 9.0; La Jolla, CA, United States) and R (version 4.2.1). The data are presented as mean ± standard deviation.
Statistical analyses were performed using a two-tailed unpaired Student’s t-test or one-way analysis of variance. Statistical differences are presented as P < 0.05, P < 0.01, and P < 0.001. P-values < 0.05 were used to denote statistical significance.
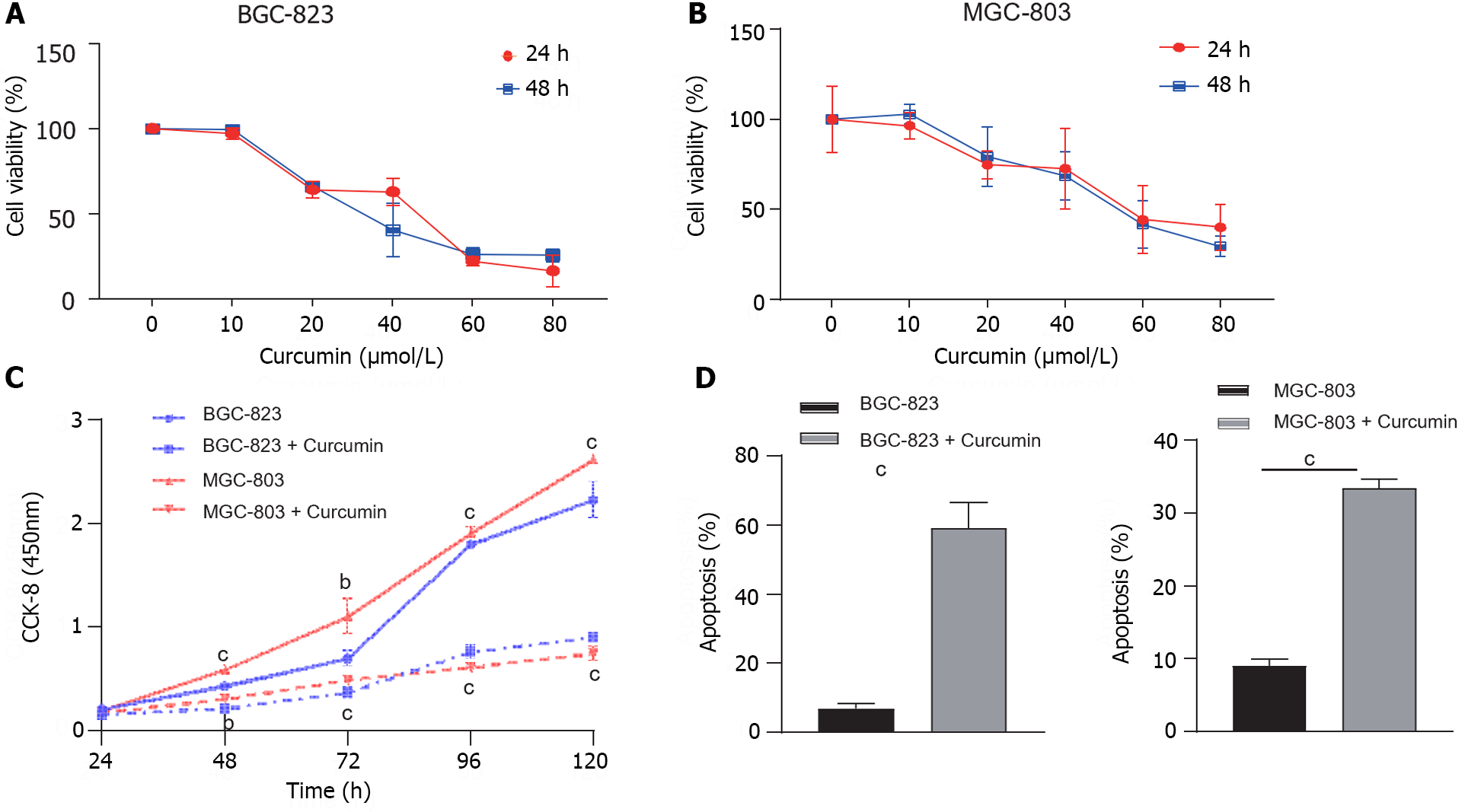
Initially, we determined the effects of curcumin on GC cell proliferation using CCK-8. BGC-823 and MGC-803 cells were treated with curcumin at concentrations of 10, 20, 40, 60, and 80 μmol/L for 24 and 48 h, respectively. Cell viability was dose-dependently reduced by curcumin treatment at various concentrations (Figure 1A and B), and we have chosen 40 μmol/L as the concentration for curcumin treatment experiments. The 40 μmol/L curcumin treatment decreased the proliferation of BGC-823 and MGC-803 cells by approximately 50% at 48 h compared to untreated cells (Figure 1C).
To further explore the potential mechanisms through which curcumin inhibits GC cell proliferation, cell apoptosis analysis was performed on BGC-823 and MGC-803 cells. The treatment with 40 μmol/L curcumin for 24 h induced 8.7-fold and 3.7-fold increased apoptosis of BGC-823 cells and MGC-803 cells, respectively (Figure 1D). These results confirmed the aforementioned proliferation inhibition activity of curcumin.
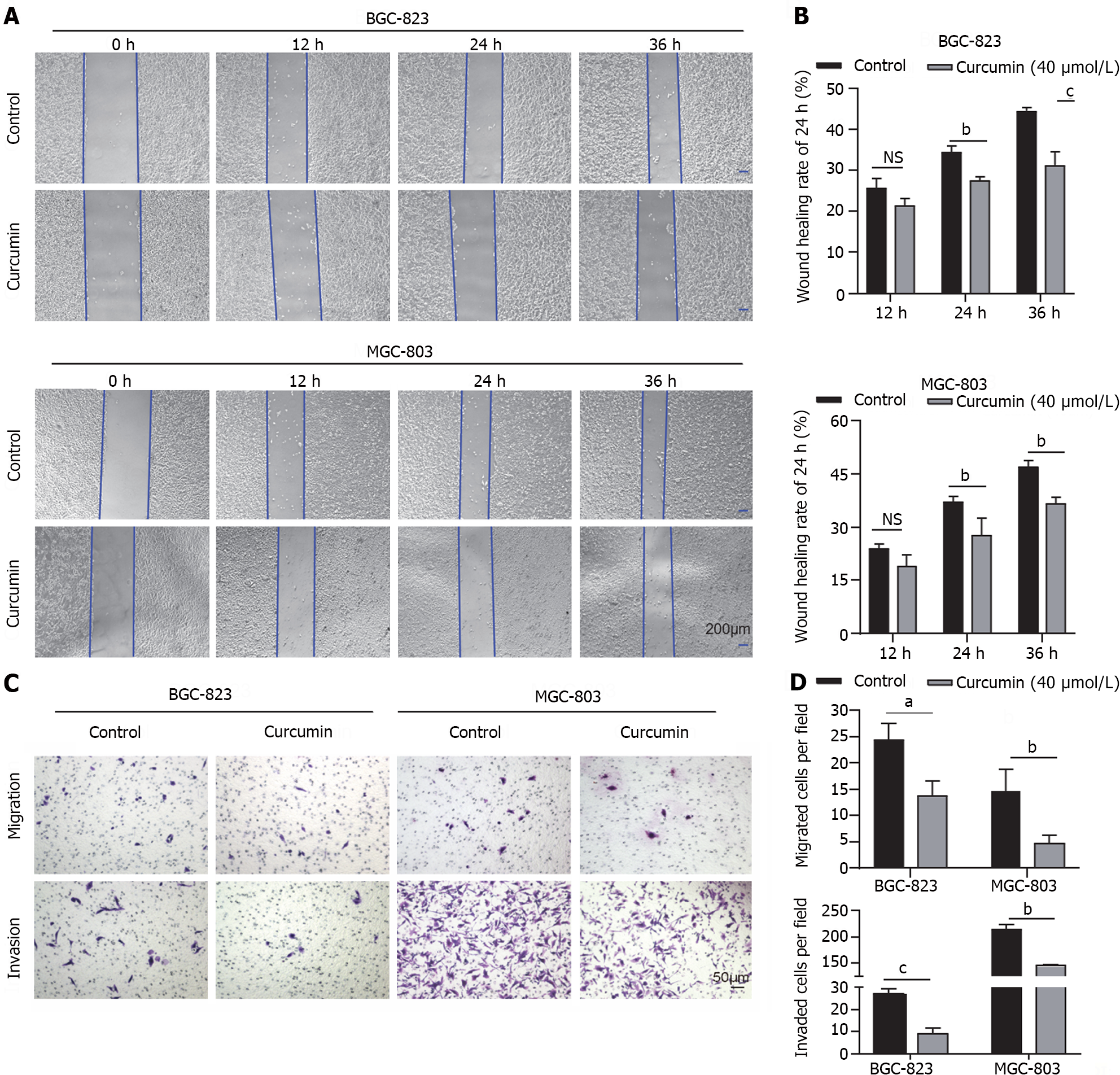
To investigate the role of curcumin in GC tumor dissemination and metastasis, wound healing and Transwell migration assays were performed. Treatment with 40 μmol/L curcumin for 24 h decreased the wound closure rate of BGC-823 cells by approximately 20% and MGC-803 cells by approximately 25% compared to untreated cells (Figure 2A and B). Furthermore, curcumin treatment decreased BGC-823 cell migration to the lower surface of chambers by 44% and MGC-803 cell migration by 68% compared to untreated cells. The Transwell invasion assay was conducted to assess the effects of curcumin on GC tumor metastasis. GC cells treated with 40 μmol/L curcumin showed decreased invasion capability compared to untreated cells. Curcumin treatment of BGC-823 cells decreased cell invasion through Matrigel by 67% compared to control cells; in MGC-803 cells, cell invasion was inhibited by approximately 32% compared to control cells (Figure 2C and D).
To confirm the role of curcumin in inducing GC cell apoptosis, we performed western blotting to measure the expression of proteins associated with cell apoptosis (Bax and Bal-2). As a master proapoptotic protein, Bax expression increased in BGC-823 and MGC-803 cells upon curcumin treatment. Consistently, the main antiapoptotic protein Bcl-2 was downregulated in curcumin-treated BGC-823 and MGC-803 cells compared to untreated cells (Figure 3A and B). Cell growth inhibition may be associated with other biological processes, such as cell cycle arrest and growth signal inactivation. In both cell lines, curcumin regulated cell cycle arrest through the cyclin D1–CDK4/6 axis because curcumin treatment decreased CDK4 and CDK6 protein levels. Furthermore, curcumin inactivated signal transduction associated with cell proliferation, as the phosphorylation of proteins, such as Akt, mTOR, and P65, was downregulated in curcumin-treated BGC-823 and MGC-803 cells compared to untreated cells (Figure 3A and B). Taken together, these results indicate that curcumin inhibits cell growth by regulating cell apoptosis, cell cycle arrest, and signal inactivation.
To explore the underlying mechanism of curcumin-mediated tumor suppression in GC, RNA-seq was performed to identify lncRNAs regulated by curcumin. Volcano plot and heatmap clustering analysis showed that 19 lncRNAs were remarkably upregulated in curcumin-treated BGC-823 cells (Figure 4A and B). We selected the top 10 lncRNAs with the most prominent fold change to confirm their expression upregulation using quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results showed that the expression of lncRNA AC022424.2 was consistently upregulated in both BGC-823 (Figure 4C) and MGC-803 (Figure 4D) cells.
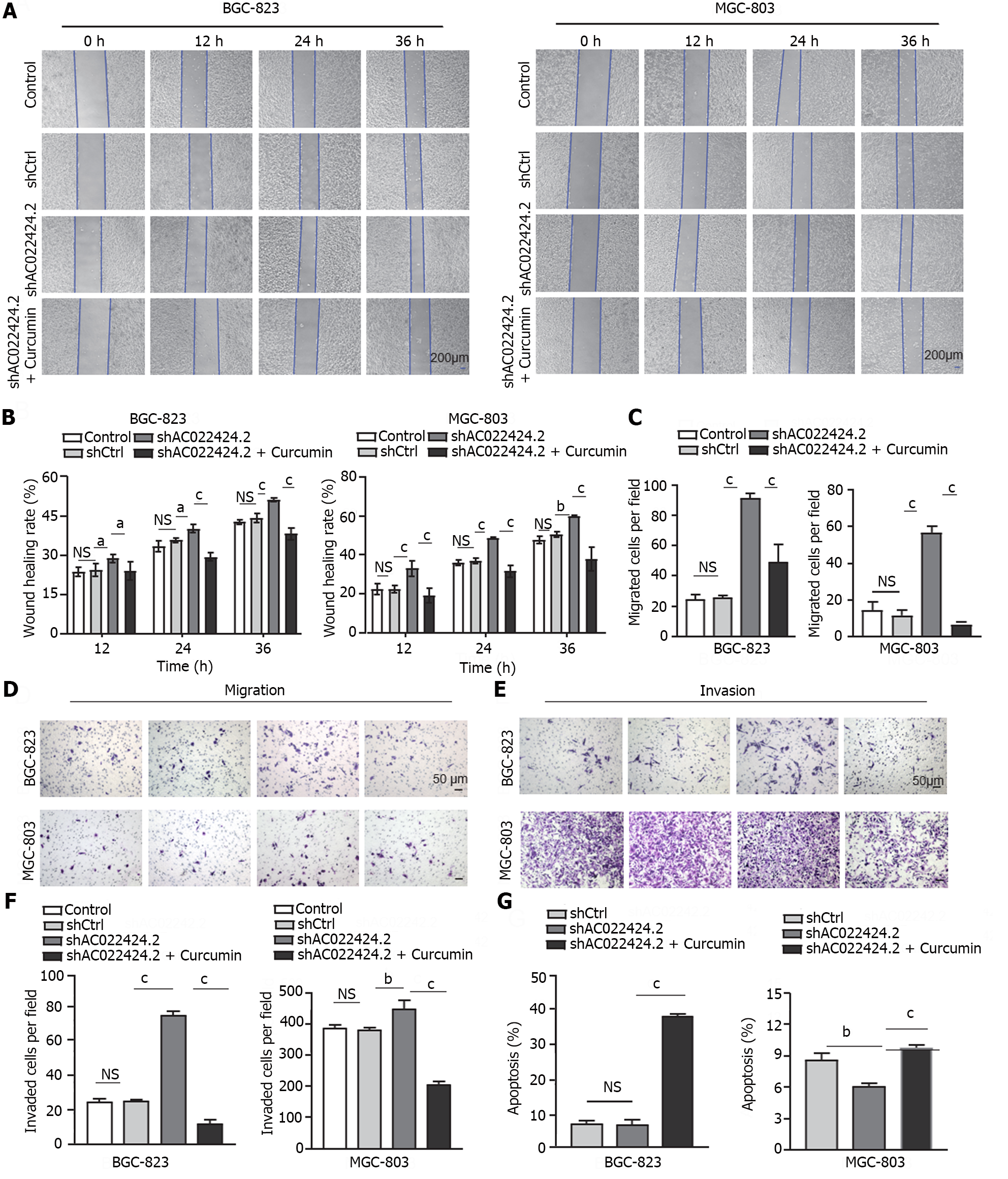
To define the function of curcumin in regulating tumor growth and metastasis through lncRNA AC022424.2, we knocked down this lncRNA using three independent shRNAs in BGC-823 and MGC-803 cells (Figure 5A and B). The knockdown efficacy was analyzed, and shAC022424.2-1 was chosen for subsequent experiments because it showed the best knockdown efficacy with a 60.16% decrease in BGC-823 cells and a 74.6% decrease in MGC-803 cells (Figure 5A and B). Meanwhile, the CCK-8 assay showed that lncRNA AC022424.2 knockdown significantly promoted cell proliferation (Figure 5C and D), wound healing, migration, and invasion (Figure 6A-F) in BGC-823 and MGC-803 cells; however, the addition of curcumin inhibited these effects. Furthermore, lncRNA AC022424.2 knockdown in BCG-823 and MGC-803 cells decreased cell apoptosis (Figure 6G). Consistently, curcumin induced more apoptosis in lncRNA AC022424.2-knockdown BCG-823 and MGC-803 cells (Figure 6G).
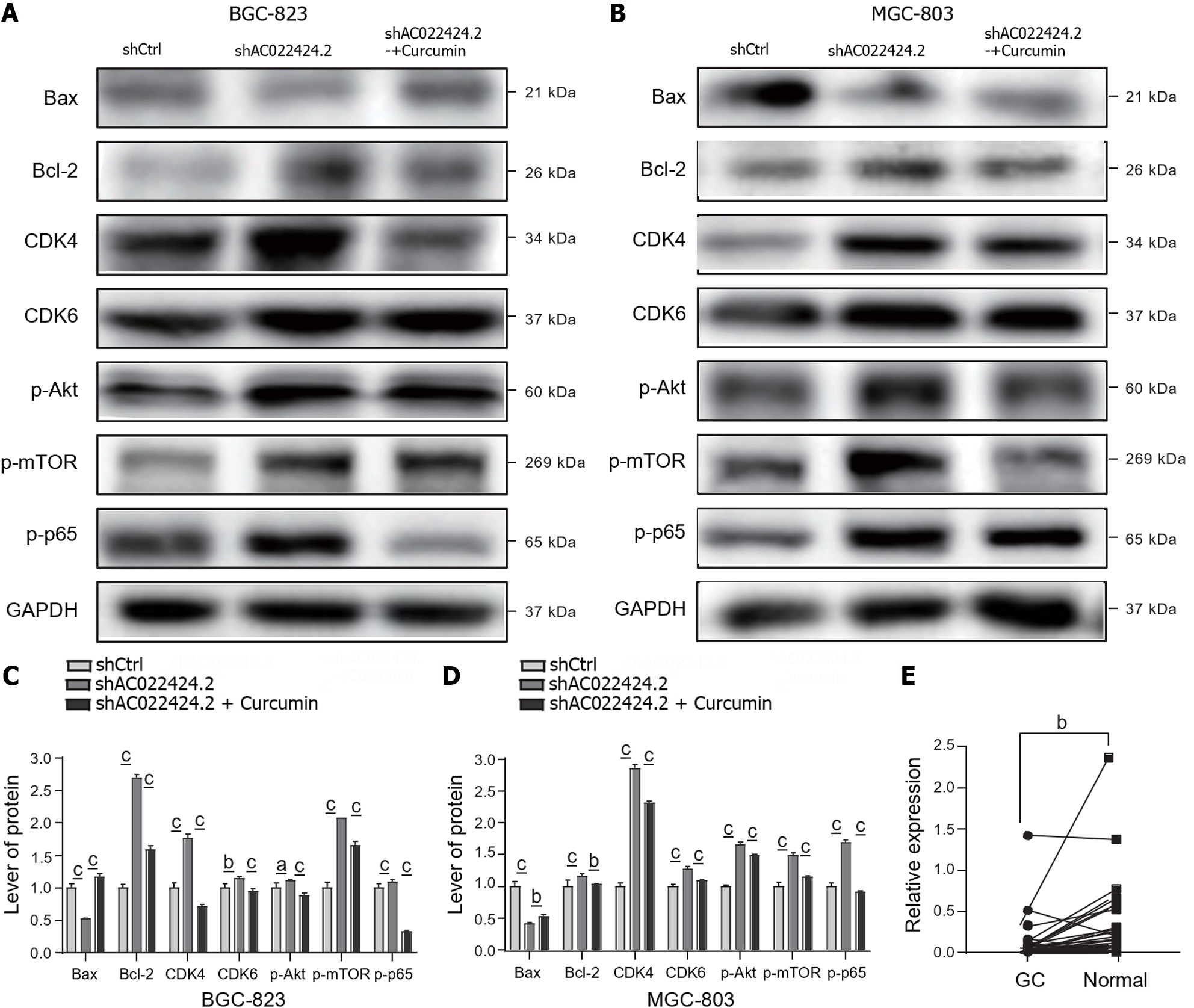
To further understand the detailed mechanisms of curcumin by regulating lncRNA AC022424.2, we performed western blotting to observe the expression of the apoptosis-associated proteins Bax and Bcl-2 and the cell cycle-related proteins CDK4 and CDK6. As shown in Figure 7A-D, lncRNA AC022424.2 knockdown decreased the expression of the proapoptotic protein Bax but augmented the expression of the antiapoptotic protein Bcl-2. Curcumin again reversed this effect. However, the cell cycle-associated proteins CDK4 and CDK6 were upregulated upon lncRNA AC022424.2 knockdown, indicating that cell cycle arrest is not the main mechanism through which curcumin regulate GC cell growth via lncRNA AC022424.2. Furthermore, lncRNA AC022424.2 knockdown activated the Akt, mTOR, and P65 signaling pathways as the phosphorylation of these three proteins was upregulated in both BGC-823 and MGC-803 cells, and curcumin treatment reduced the phosphorylation of these proteins (Figure 7A-D).
We collected 51 gastric tissues from patients with GC to investigate the clinical significance of lncRNA AC022424.2. The qRT-PCR results indicated that the expression of lncRNA AC022424.2 was significantly decreased in GC lesions compared with that in the adjacent normal gastric mucosa (Figure 7E). Furthermore, correlation analysis revealed that lower lncRNA AC022424.2 expression was significantly related to poor lymph metastasis (Table 1).
Numerous studies have substantiated the antitumor properties of curcumin, whether used in isolation or in conjunction with other pharmaceutical agents. Research has shown that curcumin can diminish H19 expression while augmenting P53 in GC cells, thereby manifesting antiproliferative effects[16]. Our experimental findings corroborate that curcumin markedly curtails the proliferation of GC cells, including AGS, MGC-803, and BGC-823 cells. This inhibitory effect intensifies with time and concentration, achieving discernible inhibition of GC cell proliferation within 48 h, corroborating previously documented literature. Moreover, our experiments revealed that curcumin fosters apoptosis in GC cells, impedes their migratory potential, and suppresses their invasive capabilities. Using sequencing technology, we identified the differentially expressed lncRNA AC022424.2 in GC cells after curcumin treatment. To investigate whether lncRNA AC022424.2 is implicated in the proapoptotic, antiproliferative, antimigratory, and anti-invasive effects of curcumin on GC cells, we constructed gene-silencing plasmid vectors. Subsequently, we transfected these vectors into MGC-803 and BGC-823 cells and conducted functional experiments that revealed that the attenuation of lncRNA AC022424.2 expression in GC cells resulted in reduced apoptosis and heightened proliferation, migration, and invasion abilities. This implies that lncRNA AC022424.2 plays a pivotal role in GC initiation and advancement.
Cancer represents an amalgamation of genetic alterations, and evading cell death serves as a fundamental change that precipitates this transformation. The literature has reported that elevated expression levels of antiapoptotic proteins, such as Bcl-2 and Bcl-xL, are associated with cisplatin resistance and tumor relapse[17,18]. Conversely, mutations in the proapoptotic gene Bax have surfaced in colorectal cancer, leading to resistance to anticancer therapies[19]. To comprehensively grasp the relationship between curcumin and apoptosis in GC cells, we used western blotting to confirm that curcumin accentuates the expression of the apoptosis-related protein Bax while diminishing the expression of Bcl-2 in MGC-803 cells. Simultaneously, the expression of the cell cycle-related proteins CDK4 and CDK6 was downregulated. Consequently, curcumin may modulate apoptosis in GC cells by recalibrating the equilibrium between proapoptotic and antiapoptotic proteins, conceivably constituting one of the underlying mechanisms driving the anti-GC effects of curcumin. Because cell proliferation is intimately correlated with tumor development, erroneous regulation of cell cycle kinases (CDKs) can lead to unbridled cell proliferation and genomic instability. CDKs are serine/threonine kinases whose catalytic activity is modulated by interactions with cell cycle proteins (cyclins) and CDK inhibitors (CKIs). This triumvirate’s concerted effort is indispensable for ensuring the orderly progression of the cell cycle[20]. Our empirical findings indicate that curcumin conceivably impedes the proliferation of GC cells by quashing the expression of CDK4 and CDK6 proteins, thereby inhibiting the cell cycle.
The PI3K/AKT/mTOR pathway assumes multiple roles in cancer progression[21]. Western blotting from our research indicates that curcumin downregulates the expression of p-AKT, p-mTOR, and p-P65. Inhibition of the PI3K/AKT/mTOR pathway elicits apoptosis and curbs proliferation. Previous investigations have confirmed the activation of the PI3K/AKT/mTOR pathway in GC, with its involvement in modulating apoptosis, autophagy, epithelial–mesenchymal transition (EMT), chemotherapy resistance, and metastasis across a spectrum of malignancies, including GC[22]. Furthermore, this pathway plays a pivotal role in bolstering cell survival by curbing apoptosis-related genes, such as Bcl-2, and propelling the activation of antiapoptotic proteins, such as NF-κB. NF-κB, which is a pivotal nuclear transcription factor, participates in inflammation, immune responses, and the orchestration of apoptosis and stress responses. Notably, apoptosis-related proteins, such as Bcl-2 and Bax, serve as targets of P65, suggesting that curcumin finetunes apoptosis, proliferation, migration, and invasion of GC cells through the PI3K/AKT/mTOR and NF-κB signaling pathways[23,24].
LncRNAs have emerged as pivotal biomarkers for early cancer screening, diagnosis, prognosis, and drug responsiveness. Recent investigations have highlighted the lncRNA MNX1-as1 (MNX1 antisense RNA 1), which registers a significant upsurge in GC tissues and is correlated with unfavorable prognoses in patients with GC[25]. With several lncRNAs potentially contributing to cancer transitions and sustenance, they hold substantial clinical significance. Our antecedent studies have attested to curcumin’s ability to induce apoptosis in GC cells while stifling their proliferation, migration, and invasion. To gain further insights into whether lncRNAs are instrumental in curcumin’s facilitation of apoptosis and impediment of proliferation, migration, and invasion of GC cells, we sequenced curcumin-treated GC cell lines and their untreated counterparts and detected 40 conspicuously differentially expressed lncRNAs, including 19 upregulated and 21 downregulated lncRNAs. We validated these outcomes using RT-PCR and identified the pivotal lncRNA AC022424.2 for subsequent cellular functional assays. Positioned on chromosome 5 and characterized by two transcripts (NR_136935.1 and ENST00000514848), lncRNA AC022424.2 displayed an approximate 3.26-fold surge in expression after curcumin treatment. Our searches across the PubMed and CNKI databases did not yield any literature related to AC022424.2. Building upon our earlier experimental findings, we postulated that curcumin may govern apoptosis, proliferation, migration, and invasion in GC through lncRNA AC022424.2. To validate our hypothesis, we conducted loss-of-function experiments on lncRNA AC022424.2. These experiments revealed the attenuation of lncRNA AC022424.2 expression through gene-silencing plasmid vectors and subsequent transfection into GC MGC-803 and BGC-823 cells. Our observations illuminated the capability of curcumin to amplify lncRNA AC022424.2 expression in GC cells. Upon repressing the expression of lncRNA AC022424.2, we noted a reduction in apoptosis, accompanied by an increase in proliferation, migration, and invasion of GC cells. This was concomitant with a decrease in the expression of the proapoptotic protein Bax, increased expression of in the antiapoptotic protein Bcl-2, and augmented expression of CDK4, CDK6, and p-mTOR. Notably, the introduction of curcumin following lncRNA AC022424.2 repression redressed these effects. These findings allude to curcumin’s probable regulation of apoptosis and proliferation in GC BGC823 and MGC803 cells through lncRNA AC022424.2, potentially via the PI3K/AKT/mTOR pathway. Xu’s research confirmed the role of lncRNA LINC01021 (also recognized as PURPL, a regulator of P53 levels) in downregulating basal P53 expression by interacting with MYBBP1A (a protein that binds to and activates P53). Da0324, a curcumin analog, has demonstrated the capacity to dampen LINC01021 expression in GC. The inhibition of LINC01021 by Da0324 curtailed GC proliferation, EMT, invasion, and spurred apoptosis in vitro. Targeted silencing of LINC01021 constrained the in vivo growth of GC xenografts[26]. The balance between proapoptotic proteins (e.g., BAX and BAK) and antiapoptotic proteins (e.g., Bcl-2 and Bcl-xL) in the regulation of the mitochondrial apoptosis pathway serves as a critical determinant, and disruption of this equilibrium frequently fosters cancer progression and chemotherapy resistance. In GC, Wang et al[27] reported that cisplatin-resistant associated lncRNA acts as a competing endogenous RNA by sponging endogenous miR-505. This upregulated the expression of the tumor suppressor CYLD, hampered AKT activation, and amplified the sensitivity of GC cells to DDP (cisplatin)[15,27]. Several investigations have revealed the activation mechanisms of the LINC1410-miR-532-5p-NCF2-NF-κB feedback loop in GC. LINC1410 and miR-532-5p may serve as prospective therapeutic targets for GC[28].
Considering gene chip technology and high-throughput sequencing’s widespread adoption, the identification of aberrantly expressed lncRNAs across diverse cancer types has increased. However, deciphering the roles of lncRNAs in cancer remains an intricate challenge. The mere observation of lncRNA dysregulation in tumor samples does not necessarily signify their causative or tumor-suppressive functions. Bioinformatics techniques have been instrumental in predicting the functions of lncRNAs by scrutinizing their common expression patterns across diverse cell and tissue types with protein-coding genes. Through these methods, several lncRNAs associated with pivotal cancer pathways, such as prostate cancer antigen 3 (also known as DD3) and prostate cancer gene expression marker 1, have been identified, initially owing to their disparate expression in prostate tumors vs healthy tissues[13,29]. Over the past decade, we have leveraged The Cancer Genome Atlas database and bioinformatics analyses to pinpoint lncRNAs that exhibited aberrant expression patterns in GC. These lncRNAs actively participate in multiple tumor signaling pathways, including Notch, mTOR, NF-κB, and Wnt[14]. Moreover, some investigations have underscored the correlation between lncRNA dysregulation and clinicopathological factors, such as TNM staging, prognosis, tumor size, and differentiation in GC[30]. Our preliminary experimental results suggest that the suppression of lncRNA AC022424.2 can deter GC cell apoptosis and stimulate migration and invasion. To substantiate the expression of AC022424.2 in tissues and contribute to early clinical diagnoses and GC treatment, we collected 64 GC specimens and performed a correlation analysis between the expression level of lncRNA AC022424.2 in GC tissues and clinicopathological data. These analyses revealed an association between the expression levels of lncRNA AC022424.2 and lymph node metastasis, with lower expression observed in GC tissues exhibiting lymph node metastasis, constituting a statistically significant difference. The panorama of lncRNAs’ roles in cancer is increasing, establishing their undeniable clinical significance. Prior research has validated lncRNAs as increasingly indispensable biomarkers for early cancer screening, diagnosis, prognosis, and drug responsiveness[31-34]. Furthermore, lncRNAs regulate chemotherapy resistance by orchestrating apoptosis, EMT, cancer stemness, autophagy, multiple drug resistance-related gene expression, and epigenetic modifications, among other signaling pathways[35]. Thus, targeting lncRNAs is a promising strategy for increasing chemotherapy sensitivity and efficacy in GC. Notwithstanding the remarkable strides in comprehending lncRNA structure and function since their discovery, the domain of lncRNAs remains in its nascent stages, with considerable terrain left to explore.
Curcumin has potential anticancer effects on gastric cancer cells by regulating lncRNA AC022424.2. This lncRNA plays a significant role in cancer cell behavior and may have clinical implications in diagnosis and prognosis evaluation. The results of this study enhance our understanding of gastric cancer development and precision treatment.
Tumor metastasis, the high heterogeneity of cancer cells, and chemotherapy resistance are primary factors that contribute to the poor overall prognosis of gastric cancer (GC). However, its role in GC treatment is constrained by adverse drug reactions and drug resistance. In the realm of exploring alternative cancer treatments, natural compounds with anticancer properties, such as curcumin, are garnering attention because of their low adverse effects. Although the anticancer effects of curcumin have been extensively studied, its specific mechanisms of action remain incompletely understood. This study employs long non-coding RNA (lncRNA) sequencing to screen for differentially expressed lncRNAs after curcumin treatment. Further investigation will explore the relationship between the ability of curcumin to inhibit GC cells and lncRNA AC022424.2.
Recent studies have highlighted the association between abnormal lncRNA expression and various aspects of gastric cancer, including early diagnosis, clinical staging, metastasis, drug sensitivity, and prognosis. A comprehensive understanding of how lncRNAs intricately influence gastric cancer development can offer new perspectives for precision treatment and personalized management of gastric cancer patients. This study aims to uncover the potential of curcumin in suppressing the malignant behavior of gastric cancer cells by upregulating specific lncRNAs and modulating the initiation and progression of gastric cancer.
The objective of this study was to identify curcumin-associated lncRNAs and examine the involvement of lncRNA AC022424.2 in the modulation of gastric cancer cell apoptosis, proliferation, and invasion upon curcumin treatment. Additionally, the validation of these findings was conducted using clinical samples.
The study used CCK-8 assays to assess curcumin's impact on gastric cancer cells, employing flow cytometry, scratch/Transwell assays, and western blotting to examine apoptosis, migration, invasion, and protein expression changes. LncRNA sequencing and quantitative reverse transcription polymerase chain reaction (qRT-PCR) validated differential expression before/after curcumin treatment in BGC-823 and MGC-803 cells. AC022424.2-1 knockdown cells were generated to scrutinize lncRNA AC022424.2's effects on apoptosis, proliferation, migration, and invasion, with western blotting assessing pathway-related protein changes. RT-PCR measured lncRNA AC022424.2 expression in clinical gastric cancer tissues, correlating with pathological characteristics.
Curcumin induced gastric cancer cell apoptosis and inhibited proliferation, migration, and invasion in a dose- and time-dependent manner. Following curcumin treatment, lncRNA AC022424.2 was upregulated, and its knockdown intensified cancer cell aggressiveness. The impact of lncRNA AC022424.2 on cancer cells may involve the PI3K/Akt/mTOR and NF-κB signaling pathways. The downregulation of lncRNA AC022424.2 was associated with lymph node metastasis, suggesting its potential as a diagnostic and prognostic marker.
Curcumin has potential anticancer effects on gastric cancer cells by regulating lncRNA AC022424.2. This lncRNA plays a significant role in cancer cell behavior and may have clinical implications in diagnosis and prognosis evaluation. The results of this study enhance our understanding of gastric cancer development and precision treatment.
The subsequent validation of these experimental results in clinical tissue specimens and exploration of specific mechanisms will provide more theoretical support for elucidating the role of lncRNAs in the occurrence and development of GC and identifying precise therapeutic targets.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hori T, Japan S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Cai YX
| 1. | Iida M, Ikeda F, Ninomiya T, Yonemoto K, Doi Y, Hata J, Matsumoto T, Iida M, Kiyohara Y. White blood cell count and risk of gastric cancer incidence in a general Japanese population: the Hisayama study. Am J Epidemiol. 2012;175:504-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 432] [Cited by in F6Publishing: 682] [Article Influence: 170.5] [Reference Citation Analysis (1)] |
| 3. | Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci Rep. 2020;10:1504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 161] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 4. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 756] [Cited by in F6Publishing: 568] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 5. | Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 2018;21:249-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 6. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 7. | Barlebo Ahlborn L, Østrup O. Toward liquid biopsies in cancer treatment: application of circulating tumor DNA. APMIS. 2019;127:329-336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Stahl P, Seeschaaf C, Lebok P, Kutup A, Bockhorn M, Izbicki JR, Bokemeyer C, Simon R, Sauter G, Marx AH. Heterogeneity of amplification of HER2, EGFR, CCND1 and MYC in gastric cancer. BMC Gastroenterol. 2015;15:7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 9. | He Z, Xu JG. Evaluation of the efficiency and safety of combined chemotherapy and molecular-targeted therapy in the treatment of advanced gastric cancer: A protocol for systematic review and meta-analysis. Medicine (Baltimore). 2021;100:e27557. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 10. | Yang JM, Sun Y, Wang M, Zhang XL, Zhang SJ, Gao YS, Chen L, Wu MY, Zhou L, Zhou YM, Wang Y, Zheng FJ, Li YH. Regulatory effect of a Chinese herbal medicine formula on non-alcoholic fatty liver disease. World J Gastroenterol. 2019;25:5105-5119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 46] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 11. | Kuttan R, Bhanumathy P, Nirmala K, George MC. Potential anticancer activity of turmeric (Curcuma longa). Cancer Lett. 1985;29:197-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 375] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 12. | Sintara K, Thong-Ngam D, Patumraj S, Klaikeaw N. Curcumin attenuates gastric cancer induced by N-methyl-N-nitrosourea and saturated sodium chloride in rats. J Biomed Biotechnol. 2012;2012:915380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Li H, Yu B, Li J, Su L, Yan M, Zhang J, Li C, Zhu Z, Liu B. Characterization of differentially expressed genes involved in pathways associated with gastric cancer. PLoS One. 2015;10:e0125013. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 15. | Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, Liu J, Xu Y, Shen Y, Yang M. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19:62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 261] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 16. | Liu G, Xiang T, Wu QF, Wang WX. Curcumin suppresses the proliferation of gastric cancer cells by downregulating H19. Oncol Lett. 2016;12:5156-5162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Han JY, Hong EK, Choi BG, Park JN, Kim KW, Kang JH, Jin JY, Park SY, Hong YS, Lee KS. Death receptor 5 and Bcl-2 protein expression as predictors of tumor response to gemcitabine and cisplatin in patients with advanced non-small-cell lung cancer. Med Oncol. 2003;20:355-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Michaud WA, Nichols AC, Mroz EA, Faquin WC, Clark JR, Begum S, Westra WH, Wada H, Busse PM, Ellisen LW, Rocco JW. Bcl-2 blocks cisplatin-induced apoptosis and predicts poor outcome following chemoradiation treatment in advanced oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2009;15:1645-1654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | Yang D, Chen MB, Wang LQ, Yang L, Liu CY, Lu PH. Bcl-2 expression predicts sensitivity to chemotherapy in breast cancer: a systematic review and meta-analysis. J Exp Clin Cancer Res. 2013;32:105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1020] [Cited by in F6Publishing: 988] [Article Influence: 43.0] [Reference Citation Analysis (1)] |
| 21. | Jafari M, Ghadami E, Dadkhah T, Akhavan-Niaki H. PI3k/AKT signaling pathway: Erythropoiesis and beyond. J Cell Physiol. 2019;234:2373-2385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 22. | Lee HJ, Venkatarame Gowda Saralamma V, Kim SM, Ha SE, Raha S, Lee WS, Kim EH, Lee SJ, Heo JD, Kim GS. Pectolinarigenin Induced Cell Cycle Arrest, Autophagy, and Apoptosis in Gastric Cancer Cell via PI3K/AKT/mTOR Signaling Pathway. Nutrients. 2018;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 23. | Erratum for Khorasani et al., The effect of omeprazole on asthmatic adolescents with gastroesophageal reflux disease. Allergy Asthma Proc. 2008;29:675. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 24. | Li C, Liang G, Yang S, Sui J, Yao W, Shen X, Zhang Y, Peng H, Hong W, Xu S, Wu W, Ye Y, Zhang Z, Zhang W, Yin L, Pu Y. Dysregulated lncRNA-UCA1 contributes to the progression of gastric cancer through regulation of the PI3K-Akt-mTOR signaling pathway. Oncotarget. 2017;8:93476-93491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 25. | Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang H, Tian S, Xu T, Shu Y. TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol Cancer. 2020;19:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 26. | Xu F, Ji Z, He L, Chen M, Chen H, Feng Q, Dong B, Yang X, Jiang L, Jin R. Downregulation of LINC01021 by curcumin analog Da0324 inhibits gastric cancer progression through activation of P53. Am J Transl Res. 2020;12:3429-3444. [PubMed] [Cited in This Article: ] |
| 27. | Wang Z, Wang Q, Xu G, Meng N, Huang X, Jiang Z, Chen C, Zhang Y, Chen J, Li A, Li N, Zou X, Zhou J, Ding Q, Wang S. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020;17:1576-1589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 28. | Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW, Gao Y, Xu Y, Chen C, Zheng ZS, Weng HW, Ye S, Kuang M, Xie D, Peng S. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660-2675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 29. | Jung M, Xu C, Spethmann J, Johannsen M, Deger S, Stephan C, Loening SA, Jung K. Re: Hessels D, Klein Gunnewiek JMT, van Oort I, Karthaus HFM, van Leenders GJL, van Balken B, Kiemeney LA, Witjes JA, Schalken JA. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003;44:8-16. Eur Urol. 2004;46:271-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Atala A. Re: lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. J Urol. 2014;191:1470-1471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 31. | Zuo Z, Hu H, Xu Q, Luo X, Peng D, Zhu K, Zhao Q, Xie Y, Ren J. BBCancer: an expression atlas of blood-based biomarkers in the early diagnosis of cancers. Nucleic Acids Res. 2020;48:D789-D796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689-4698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 34. | Sasaki N, Iwaya T, Chiba T, Fujita M, Ju Z, Endo F, Yaegashi M, Hachiya T, Sugimoto R, Sugai T, Siwak DR, Liotta LA, Lu Y, Mills GB, Nakagawa H, Nishizuka SS. Analysis of mutational and proteomic heterogeneity of gastric cancer suggests an effective pipeline to monitor post-treatment tumor burden using circulating tumor DNA. PLoS One. 2020;15:e0239966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Lin KN, Jiang YL, Zhang SG, Huang SY, Li H. Grape seed proanthocyanidin extract reverses multidrug resistance in HL-60/ADR cells via inhibition of the PI3K/Akt signaling pathway. Biomed Pharmacother. 2020;125:109885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |