Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.314
Peer-review started: October 12, 2023
First decision: October 28, 2023
Revised: December 12, 2023
Accepted: January 5, 2024
Article in press: January 5, 2024
Published online: February 15, 2024
Cyclin-dependent kinase 9 (CDK9) expression and autophagy in colorectal cancer (CRC) tissues has not been widely studied. CDK9, a key regulator of transcription, may influence the occurrence and progression of CRC. The expression of auto
To investigate the relationship between CDK9 expression and autophagy in CRC, assess differences in autophagy between left and right colon cancer, and analyze the associations of autophagy-related genes with clinical features and prognosis.
We collected tumor tissues and paracarcinoma tissues from colon cancer patients with liver metastasis to observe the level of autophagy in tissues with high levels of CDK9 and low levels of CDK9. We also collected primary tissue from left and right colon cancer patients with liver metastasis to compare the autophagy levels and the expression of BECN1 and ABCG2 in the tumor and paracarcinoma tissues.
The incidence of autophagy and the expression of BECN1 and ABCG2 were different in left and right colon cancer, and autophagy might be involved in the occurrence of chemotherapy resistance. Further analysis of the rela
This study laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of patients with CRC.
Core Tip: Autophagy has a dual role in tumorigenesis: inhibiting tumorigenesis under normal conditions and promoting tumor growth once formed. In colon cancer patients with liver metastasis, high cyclin-dependent kinase 9 (CDK9) expression correlated with elevated autophagy levels. Autophagy incidence and expression of BECN1 and ABCG2 differed between left and right colon cancer, potentially contributing to chemotherapy resistance. High CDK9 expression was associated with poor prognosis in colorectal cancer. This study provided a foundation for investigating CDK9 and autophagy inhibitors in combination therapy to enhance tumor cell sensitivity to chemotherapy.
- Citation: Zheng L, Lu J, Kong DL. Expression of cyclin-dependent kinase 9 is positively correlated with the autophagy level in colon cancer. World J Gastrointest Oncol 2024; 16(2): 314-330
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/314.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.314
Colorectal cancer (CRC) is one of the most common malignant tumors in China, and its incidence rate is increasing every year. The latest research has confirmed that there are many differences between left-sided colon cancer and right-sided colon cancer in embryologic origin, pathogenesis, histopathology, epidemiology, epigenetics, clinical manifestations, and prognosis[1]. Some scholars postulate that left-sided colon cancer and right-sided colon cancer are two distinct diseases[2]. Therefore, the comparative study of left-sided and right-sided colon cancer is helpful to understand pathogenesis and the mechanism of progression.
Microscopically, the occurrence and development of tumors are mainly characterized by unlimited abnormal proliferation of cells. The proliferation and differentiation of cells in multicellular organisms[3,4] are regulated by the cell cycle. The main causes of tumor occurrence are cell cycle disorder and increased resistance to apoptosis. Cyclin-dependent kinases (CDKs) promote DNA synthesis and mitosis[5,6] through phosphorylation of key substrates and participate in cell cycle regulation. Abnormal activation of CDKs leads to the imbalance of the cell cycle, uncontrollable cell proliferation, and the occurrence and development of malignant tumors[7]. For example, the CDK family includes 20 kinases that contribute to cell deterioration by promoting proliferation, migration, invasion, and anti-apoptosis of cancer cells[8].
CDK9 is a key regulator of transcription and is used to continuously produce short-lived proteins to maintain the survival of cancer cells[9]. As a part of a functionally diverse group of enzymes responsible for cell cycle control and progression, CDK9 associates primarily with cyclin T1 and forms the positive transcription elongation factor b (p-TEFb) complex responsible for the regulation of transcription elongation and mRNA maturation. CDK9 can influence almost all cellular functions. Many studies have shown that the dysfunction of CDK9 signaling is obvious in various hematologic malignancies and solid tumor cell lines, and the inhibition of CDK9 can lead to growth suppression and apoptosis of cancer cells[10,11]. Due to the extremely critical role of CDK9 in cancer cells, inhibiting its functions has been a research hotspot, with more and more studies focusing on small, molecular inhibitors, some of which are presently in clinical trials. The search for newer CDK9 inhibitors with higher specificity and lower toxicities continues[12]. Targeting CDK9 with wogonin and related natural flavones could enhance the anticancer effect of the BCL-2 family inhibitor ABT-263[13].
Macroautophagy, commonly known as autophagy, is the main form of cell autophagy. It sequesters proteins, lipids, or organelles that need to be degraded with a bimolecular membrane structure, forming autophagosomes. Autophagosomes can be integrated with lysosomes to form an autolysosome, which degrades the encapsulated cellular components through proteolytic enzymes and lipases in lysosomes and then recycles or excretes them. Autophagy in tumor cells is a double-edged sword. Autophagy can eliminate abnormally folded proteins and dysfunctional organelles, inhibit the stress response of cells, and ultimately prevent genome damage, thus inhibiting the occurrence of cancer[14]. On the other hand, tumor cells can utilize autophagy to promote their survival under conditions of nutritional deficiency and hypoxia[15].
The relationship between autophagy and the progression of tumorigenesis is complicated. The promotion or suppression of autophagy depends on the tumor type and environment[16]. The number of autophagosomes observed by transmission electron microscope (TEM) in cancer tissues can reflect the level of autophagy[17]. In vitro and in vivo data support the hypothesis that autophagy could enhance cancer cell drug resistance to chemotherapy, and inhibition of autophagy may increase the sensitivity of cancer cells to chemotherapy[18,19].
Evidence suggests that BECN1 is a tumor suppressor. BECN1 binds and activates VP34 through a conserved domain to induce autophagy[20]. Through the positive regulators of BECN1, UVRAG, and BIF-1, the combination of BECN1 and VP34 is enhanced, leading to an increase in autophagy and resulting in tumor inhibition[21]. Overexpression of anti-apoptotic protein BCL-2 promotes the survival of breast cancer, prostate cancer, small cell lung cancer, non-small cell lung cancer, and leukemia[22,23]. The interaction between BCL-2 and BECN1 could be exploited to prevent autophagy[24].
Mitochondrial autophagy (also known as mitophagy) is based on selective degradation dysfunction or damaged mitochondria and plays an important role in maintaining high-quality mitochondria[25]. Regulation of this process is critical for maintaining cellular homeostasis and has been closely associated with acquired drug resistance. CDK9 may be closely associated with the regulation of mitochondrial function and mitophagy as well. It was reported that inhibition of CDK9 blocked PINK1-PRKN-mediated mitophagy in hepatocellular carcinoma by interrupting mitophagy initiation[26]. However, the relationship between autophagy and CDK9 in CRC has not been fully investigated. Therefore, elucidating the interaction between autophagy and CDK9 would be helpful for improving the efficacy of anticancer drugs in the clinical treatment of CRC.
The ATP-binding cassette transmembrane transporter superfamily is involved in the transmembrane transport of anticancer drugs, which decreases the toxicity of anticancer drugs for tumor cells. The ABCG2 transporter mediates multidrug resistance and regulates autophagy. Under normal growth conditions, cells with high levels of ABCG2 have a higher basic autophagy rate than cells with low levels of ABCG2, which could be associated with the response to stressors[27]. For example, high expression of ABCG2 was associated with chemotherapy resistance and poor prognosis in patients with osteosarcoma[28]. In childhood leukemia and chronic lymphocytic leukemia, drug resistance to adriamycin increased after overexpression of ABCG2[29].
In this study, the relationship between the expression of CDK9 and the incidence of autophagy in CRC was determined. We also compared the incidence of autophagy and prognosis with ABCG2 expression in left and right colon cancers. We found that the expression of CDK9 was positively correlated with autophagy in CRC. The incidence rate of autophagy and expression of ABCG2 in patients with liver metastasis between left and right colon cancers was different, and autophagy may be involved in the resistance to chemotherapy. We further analyzed the relationship among the expression of autophagy-related genes CDK9, ABCG2, and BECN1, the clinical features, and the prognosis of CRC.
Public gene expression data and corresponding clinical annotations for transcriptome analysis were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/; Date: March 31, 2021; Version: V29.0). RNAseq data in level 3 HTSeq FPKM format in TCGA-COAD and TCGA-READ were used. The RNAseq data in FPKM format was converted into TPM format, and log2 conversion was performed. The clinicopathological features of public data are shown in Supplementary Table 1.
Nineteen groups of fresh tumor and paracarcinoma colon tissue derived from 19 patients were collected. Twelve patients were diagnosed with left colon cancer, and seven patients were diagnosed with right colon cancer. All patients had liver metastasis. Twenty-four groups of fresh tumor tissue and paired tissue from 24 patients were collected. Fifteen patients were diagnosed with stage IV left colon cancer, and nine patients were diagnosed with right colon cancer. The basic information of the sample group is shown in Supplementary Table 2. Fresh tissues were defined as fresh surgical tissues, soybean size 1-2, and 40-50 mg. The proportion of tumor cells was greater than 40%, and the proportion of necrotic cells was not greater than 10%. Informed consent was obtained from all subjects and/or their legal guardians.
Electron microscopy samples of 24 groups of fresh tumor tissue and paracarcinoma tissue from stage IV left and right colon cancer patients were prepared according to a previously published protocol[30]. Nineteen groups of fresh tumor and paracarcinoma tissues derived from 19 patients with left and right colon cancer with liver metastasis were used for immunohistochemistry (IHC) based on protocols in our facility’s pathology department.
For TEM, tumor and paracarcinoma tissues from stage IV colon cancer patients were fixed in 2.5% glutaraldehyde, rinsed in 0.1 M phosphoric acid, and then immobilized with 1% osmic acid fixative. Dehydration included sequential steps in ethanol and acetone. Samples were embedded, solidified, and sliced into 70-nm sections. Staining was performed with 3% uranium acetate-lead citrate. Finally, observation and filming were carried out using the JEOL JEM-1230 TEM at 80 KV.
For IHC, paraffin section preparation involved tissue dehydration, transparency, wax immersion, block embedding, and sectioning. Dewaxing included xylene, 100% alcohol, and subsequent alcohol concentrations with 10-min intervals. Duration varied with temperature. For antigen repair, the tissues were washed, treated with 3% hydrogen peroxide for 10 min and then citric acid buffer before being microwaved for antigen exposure. Serum blocking involved washing with phosphate buffered saline, drying, serum addition, and incubation at 37 °C. The serum was diluted 10 times. Primary antibody was applied, and slides were stored at 4 °C overnight. Secondary antibody was applied after phosphate buffered saline washing. StreptAvidin-Biotin Complex solution was applied for 30 min at 37 °C. The slides were exposed to developer solution, and staining with hematoxylin was completed in 0.5-5.0 min. Dehydration included various alcohol concentrations and xylene. Slides were finally sealed and dried.
Autophagy was analyzed by flow cytometry using the CYTO-ID® Autophagy Detection kit (Enzo Life Sciences, Farmingdale, NY, United States).
Gene expression and clinical data were analyzed using R (version 3.6.3). Numerical variables were expressed as median and 25th-75th percentile. Categorical variables were expressed as numbers and percentages. The Student’s t test was used for normally distributed data, and the Mann Whitney U test was used for comparing non-normally distributed data. The χ2 test was used to compare categorical data. For paired samples that met the normality test, the paired sample t test was used. For paired samples that did not meet the normality test, the Wilcoxon signed-rank test was used.
The median gene expression level was the cutoff point, and the expression was divided into high or low expression levels. Kaplan-Meier survival analysis was applied to evaluate the overall survival rate (OS), disease-specific survival rate (DSS), and progression-free interval (PFI) of CRC at high or low expression levels. To find the factors related to OS, DSS, and PFI, we used the Cox regression model for analysis. The area under the curve (AUC) of the time-dependent receiver operating characteristic (ROC) curve was used to evaluate the accuracy of ABCG2, CDK9, and BECN1 expression levels in predicting the prognosis of patients. Autophagy pathway genes in the react database and CDK9 were imported into the string database to build protein-protein interaction (PPI) networks. CDK9 interaction genes were filtered and transferred to Cytoscape to make a PPI network. P values < 0.05 were considered significant.
TCGA-COAD and TCGA-READ analyses showed that the expression of CDK9 was increased in 50 paired CRC tissues (Figure 1, Supplementary Table 1).
We collected tumor tissues and paracarcinoma tissues from colon cancer patients with liver metastasis. We observed by TEM that tissues with a high level of CDK9 had significantly higher levels of autophagy than the tissues with a low expression of CDK9 (Figure 2). We demonstrated that the expression of CDK9 in colon cancer was positively correlated with autophagy. PPI networks of CDK9 are shown in Supplementary Figure 1. CDK9 interacts with CSNK2B, HSP90
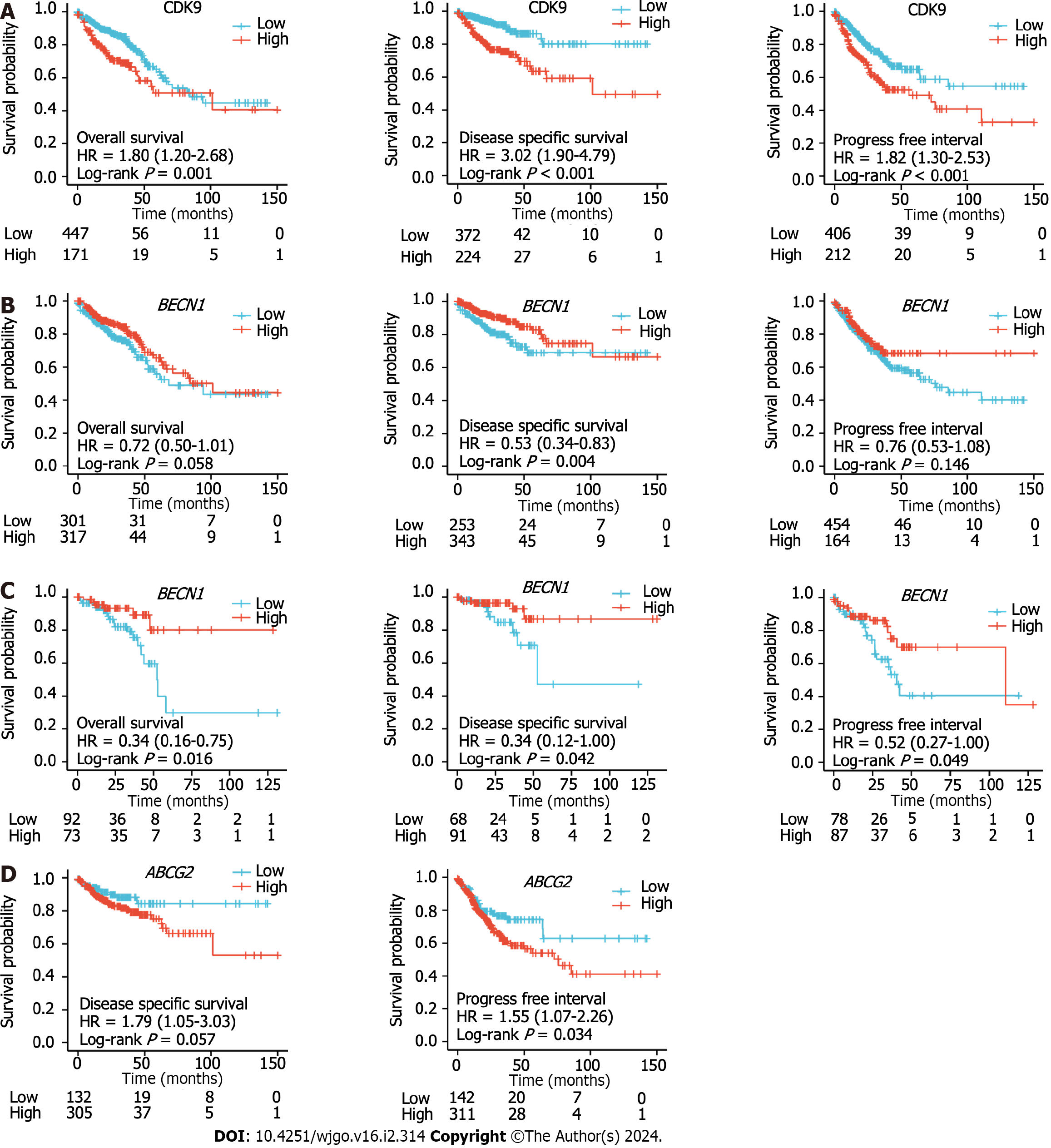
We used Cox regression analysis to determine whether CDK9 expression levels could be used to evaluate the prognosis of patients with CRC. A high expression level of CDK9 was a risk factor for the prognosis of CRC that was associated with OS and DSS (all P < 0.050; Figure 3, Tables 1 and 2). Multivariate Cox regression analysis revealed that high expression of CDK9 was an independent prognosis marker associated with OS and DSS (all P < 0.050; Figure 3, Tables 1 and 2).
| Characteristic | Category | Low expression of CDK9, n = 322 | High expression of CDK9, n = 322 | P value |
| T stage | T1 | 10 (1.6) | 10 (1.6) | 0.014 |
| T2 | 61 (9.5) | 50 (7.8) | ||
| T3 | 225 (35.1) | 211 (32.9) | ||
| T4 | 24 (3.7) | 50 (7.8) | ||
| N stage | N0 | 192 (30.0) | 176 (27.5) | 0.108 |
| N1 | 78 (12.2) | 75 (11.7) | ||
| N2 | 49 (7.7) | 70 (10.9) | ||
| M stage | M0 | 236 (41.8) | 239 (42.4) | 0.368 |
| M1 | 39 (6.9) | 50 (8.9) | ||
| Primary therapy outcome | PD | 13 (4.2) | 20 (6.4) | 0.185 |
| SD | 1 (0.3) | 4 (1.3) | ||
| PR | 10 (3.2) | 6 (1.9) | ||
| CR | 138 (44.2) | 120 (38.5) | ||
| Sex | Female | 143 (22.2) | 158 (24.5) | 0.269 |
| Male | 179 (27.8) | 164 (25.5) | ||
| CEA level | ≤ 5 | 135 (32.5) | 126 (30.4) | 0.944 |
| > 5 | 81 (19.5) | 73 (17.6) | ||
| Perineural invasion | No | 82 (34.9) | 93 (39.6) | 0.621 |
| Yes | 31 (13.2) | 29 (12.3) | ||
| Lymphatic invasion | No | 193 (33.2) | 157 (27) | 0.008 |
| Yes | 101 (17.4) | 131 (22.5) | ||
| History of colon polyps | No | 192 (34.6) | 185 (33.3) | 1.000 |
| Yes | 91 (16.4) | 87 (15.7) | ||
| Neoplasm type | Colon adenocarcinoma | 235 (36.5) | 243 (37.7) | 0.528 |
| Rectum adenocarcinoma | 87 (13.5) | 79 (12.3) | ||
| Age in yr, median (IQR) | 68.5 (60.0, 76.8) | 67.0 (57.0, 75.0) | 0.104 |
| Characteristic | Category | Low expression of ABCG2, n = 322 | High expression of ABCG2, n = 322 | P value |
| T stage | T1 | 15 (2.3) | 5 (0.8) | 0.114 |
| T2 | 54 (8.4) | 57 (8.9) | ||
| T3 | 219 (34.2) | 217 (33.9) | ||
| T4 | 33 (5.1) | 41 (6.4) | ||
| N stage | N0 | 209 (32.7) | 159 (24.8) | < 0.001 |
| N1 | 76 (11.9) | 77 (12.0) | ||
| N2 | 36 (5.6) | 83 (13.0) | ||
| M stage | M0 | 239 (42.4) | 236 (41.8) | 0.111 |
| M1 | 36 (6.4) | 53 (9.4) | ||
| Primary therapy outcome | PD | 12 (3.8) | 21 (6.7) | 0.305 |
| SD | 2 (0.6) | 3 (1.0) | ||
| PR | 10 (3.2) | 6 (1.9) | ||
| CR | 129 (41.3) | 129 (41.3) | ||
| Sex | Female | 143 (22.2) | 158 (24.5) | 0.269 |
| Male | 179 (27.8) | 164 (25.5) | ||
| CEA level | ≤ 5 | 127 (30.6) | 134 (32.3) | 0.986 |
| > 5 | 74 (17.8) | 80 (19.3) | ||
| Perineural invasion | No | 90 (38.3) | 85 (36.2) | 0.351 |
| Yes | 26 (11.1) | 34 (14.5) | ||
| Lymphatic invasion | No | 192 (33.0) | 158 (27.1) | 0.007 |
| Yes | 100 (17.2) | 132 (22.7) | ||
| History of colon polyps | No | 182 (32.8) | 195 (35.1) | 0.510 |
| Yes | 92 (16.6) | 86 (15.5) | ||
| Neoplasm type | Colon adenocarcinoma | 244 (37.9) | 234 (36.3) | 0.417 |
| Rectum adenocarcinoma | 78 (12.1) | 88 (13.7) | ||
| Age in yr, median (IQR) | 68.0 (59.0, 75.8) | 68.0 (57.3, 76.0) | 0.423 |
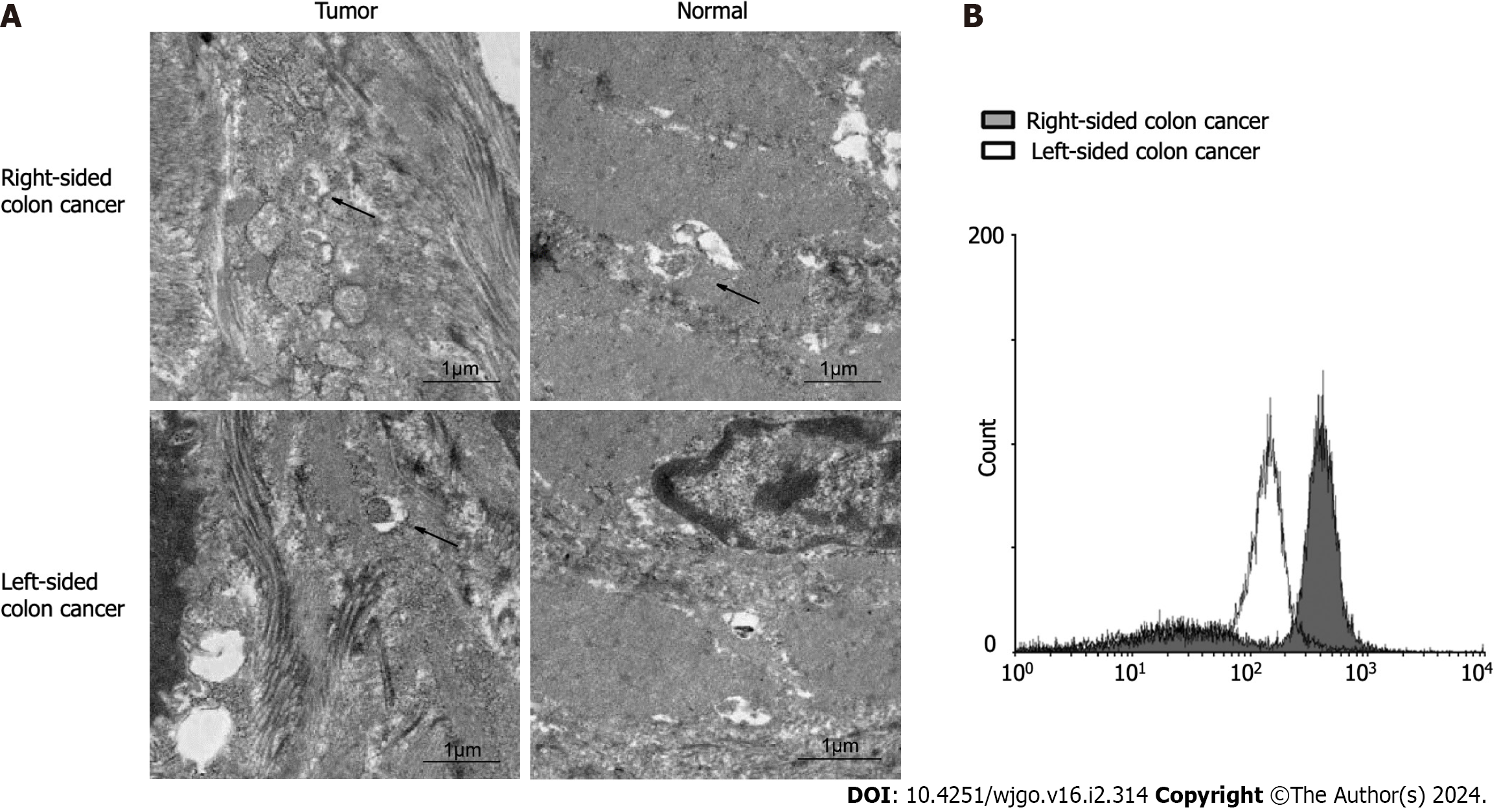
We collected 24 groups of fresh tumor tissue and paired tissue from 24 patients with stage IV left and right colon cancer. We observed autophagy with a projection electron microscope and counted the positive rate of autophagy in multiple fields. The results showed that the positive rate (8/9) of autophagy in right colon cancer was significantly higher than that in left colon cancer (5/15; χ2 = 6.993, P = 0.030) and paracarcinoma tissue (3/9; χ2 = 5.844, P = 0.016) (Figure 4A). Primary cells were isolated from the fresh tumor tissues of colon cancer patients with liver metastasis. The CYTO-ID® autophagy detection reagent was used for staining. Flow cytometry was used to detect the formation of autophagy in the primary cancer cells. The results (Figure 4B) were consistent with the observations from electron microscopy.
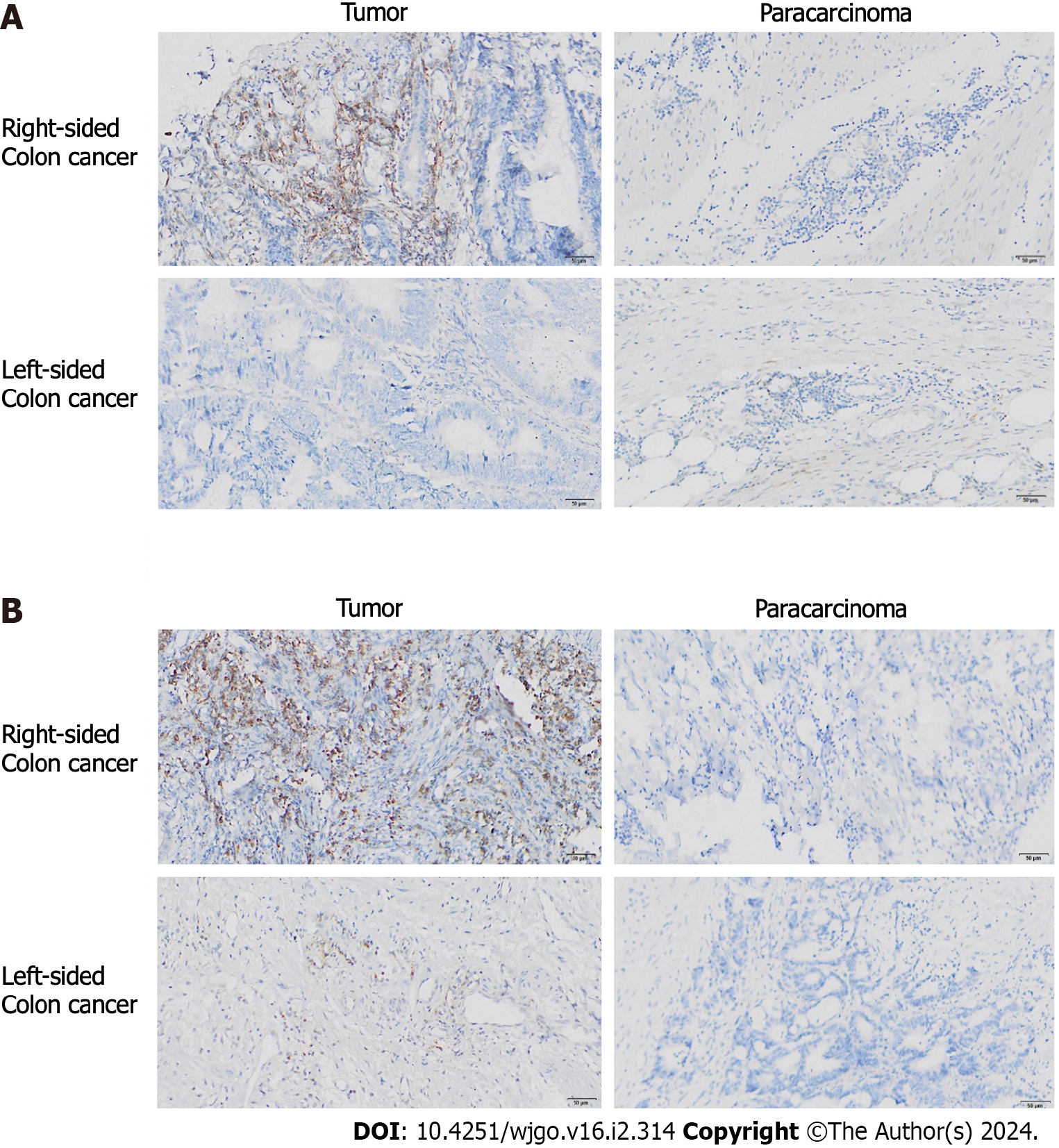
We compared the expression of BECN1 and ABCG2 between 19 groups of fresh tumor tissue and paracarcinoma tissue from colon cancer patients with liver metastasis. We also compared the expression patterns of BECN1 and ABCG2 in left and right CRC. IHC results showed that there were no significant differences in the positive probability and expression level of BECN1 between advanced colon cancer and paracarcinoma tissue. However, the positive rate of right colon cancer (72.2%, 6/7) was significantly higher than that of left colon cancer (3/12; χ2 = 6.537; P = 0.011; Figure 5A). In addition, we found that compared with the paracarcinoma tissues, the right colon cancer tissue showed a high expression of ABCG2, while the expression level of ABCG2 in the left colon cancer had no significant difference compared with the paracarcinoma tissues (Figure 5B).
These results suggest that the incidence of autophagy in the primary liver metastasis of left and right colon cancers is different, and it may be involved in the occurrence of chemotherapy resistance.
Basic information of BECN1, CDK9, and ABCG2 expression levels from 644 CRC samples in relation to clinical data were analyzed (Tables 1-3 and Supplementary Table 2). The expression levels of CDK9 were associated with tumor T stage and lymph node invasion (Table 1). The expression levels of ABCG2 were associated with tumor N-stage and lymph node invasion (Table 2). The expression levels of BECN1 were related to age, history of colon polyps, and lymph node invasion (Table 3). There was no correlation between CDK9 and age and ABCG2 and age (P = 0.821 and P = 0.388, respectively). There was a weak correlation between BECN1 and age (rho = -0.135, P = 0.001). There was no correlation between CDK9 and tumor stage and BECN1 and tumor stage (P = 0.368 and P = 0.884, respectively). There was a weak correlation between ABCG2 and tumor stage (rho = 0.105, P = 0.009).
| Characteristic | Category | Low expression of BECN1, n = 322 | High expression of BECN1, n = 322 | P value |
| T stage | T1 | 11 (1.7) | 9 (1.4) | 0.599 |
| T2 | 58 (9.0) | 53 (8.3) | ||
| T3 | 210 (32.8) | 226 (35.3) | ||
| T4 | 41 (6.4) | 33 (5.1) | ||
| N stage | N0 | 179 (28.0) | 189 (29.5) | 0.523 |
| N1 | 76 (11.9) | 77 (12.0) | ||
| N2 | 65 (10.2) | 54 (8.4) | ||
| M stage | M0 | 246 (43.6) | 229 (40.6) | 0.496 |
| M1 | 42 (7.4) | 47 (8.3) | ||
| Primary therapy outcome | PD | 16 (5.1) | 17 (5.4) | 0.775 |
| SD | 2 (0.6) | 3 (1.0) | ||
| PR | 10 (3.2) | 6 (1.9) | ||
| CR | 128 (41) | 130 (41.7) | ||
| Sex | Female | 149 (23.1) | 152 (23.6) | 0.874 |
| Male | 173 (26.9) | 170 (26.4) | ||
| CEA level | ≤ 5 | 126 (30.4) | 135 (32.5) | 0.477 |
| > 5 | 68 (16.4) | 86 (20.7) | ||
| Perineural invasion | No | 58 (24.7) | 117 (49.8) | 0.310 |
| Yes | 15 (6.4) | 45 (19.1) | ||
| Lymphatic invasion | No | 160 (27.5) | 190 (32.6) | 0.014 |
| Yes | 131 (22.5) | 101 (17.4) | ||
| History of colon polyps | No | 177 (31.9) | 200 (36) | < 0.001 |
| Yes | 113 (20.4) | 65 (11.7) | ||
| Neoplasm type | Colon adenocarcinoma | 234 (36.3) | 244 (37.9) | 0.417 |
| Rectum adenocarcinoma | 88 (13.7) | 78 (12.1) | ||
| Age in yr, median (IQR) | 69.0 (62.0, 77.0) | 65.5 (55.3, 75.0) | < 0.001 |
The Kaplan-Meier survival analysis was used to evaluate the OS, DSS, and PFI of CRC patients with high or low expression levels of BECN1, CDK9, and ABCG2. Univariate and multivariate analysis of OS, disease-free survival, and DSS for CDK9, ABCG2, and BECN1 are shown in Supplementary Table 3 and Supplementary Figure 2. The high expre
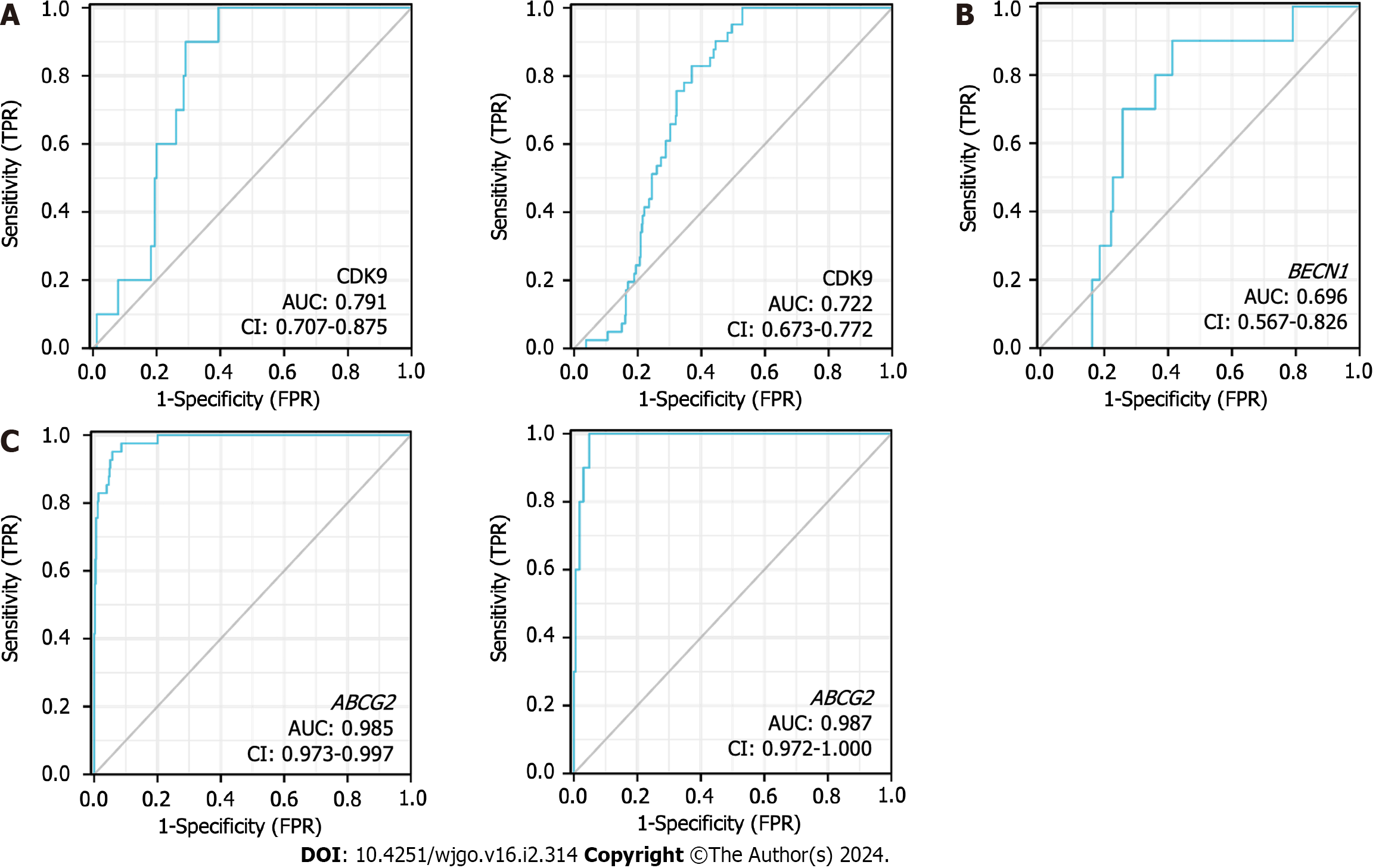
According to the expression levels of CDK9 (Figure 6A), BECN1 (Figure 6B), and ABCG2 (Figure 6C), the AUC of the time-dependent ROC curve was used to evaluate the prediction accuracy of prognosis in CRC patients. The larger the AUC value, the higher the accuracy of the prediction. ABCG2 expression levels showed the highest level of accuracy (AUC = 0.985 for rectal cancer; AUC = 0.987 for colon cancer) for the prognosis of CRC. CDK9 showed the second-highest level of accuracy for the prognosis of CRC, while BECN1 showed a lower accuracy for the prognosis of CRC.
CDK9 is the catalytic kinase of p-TEFb, which is the key to the transcription elongation of most protein-coding genes[34]. Studies have shown that CDK9 is highly expressed in CRC, breast cancer, prostate cancer, and lung cancer, and it is closely related to the malignant degree of the tumors[35]. In the early stage of tumorigenesis, autophagy serves as a survival approach and quality control mechanism to prevent the occurrence of tumors and inhibit the development of tumors. However, once the tumors are formed, autophagy can be subjected to environmental pressure, which contributes to the survival and growth of the formed tumors and promotes the invasion of the cancer by promoting metastasis[36]. Few studies on the relationship between CDK9 and autophagy have been conducted.
CDK9 inhibitors promote dephosphorylation of SIRT1 and degradation of FOXO3, which is regulated by its acetylation, mediating the transcriptional repression of FOXO3-driven BNIP3 and impairing the BNIP3-mediated stability of the PINK1 protein. Lysosomal degradation inhibitors cannot rescue mitophagy flux blocked by CDK9 inhibitors. Thus, CDK9 inhibitors inactivate the SIRT1-FOXO3-BNIP3 axis and PINK1-PRKN pathway to subsequently block mitophagy initiation in hepatocellular carcinoma. CDK9 alongside mTOR complexes modulate translation. TOR complexes have recently emerged as a potential target for autophagic regulation[37]. This study confirmed that CDK9 is highly expressed in CRC and that expression of CDK9 is positively correlated with autophagy in colon cancer.
ABCG2 plays a crucial role in drug transport and cellular defense mechanisms. In vitro experiments have demonstrated that the expression of ABCG2 can enhance cell survival and resistance to adverse conditions, such as amino acid starvation and radiation-associated cell death. These resistance mechanisms are closely associated with ABCG2-dependent autophagy, a cellular process that helps maintain cell homeostasis and repair damage[27]. The enhancement of autophagy depends on the overexpression of ABCG2[27]. The positive rate of autophagy in right colon cancer was significantly higher than that in the left colon cancer. We also found that the expression level of ABCG2 in right colon cancer was higher than that in the paracarcinoma tissue, while there was no significant difference between the expression level of ABCG2 in left colon cancer and the paracarcinoma tissue (Figure 5B). The root cause of chemotherapy resistance in breast cancer is closely related to cancer stem cell subsets, and their activation depends on paclitaxel-promoted autophagy[38]. It was found that rapamycin-induced autophagy could reduce the chemosensitivity of breast cancer cells to the XIAOPI formula[39].
High expression of the CDK9 marker represented a poor prognosis for patients with CRC. The expression CDK9 was upregulated in patients with papillary thyroid carcinoma, and it was associated with prognosis in these patients[40]. High expression of CDK9 is an independent marker for the poor prognosis in osteosarcoma[41]. CDK9 is also highly expressed in human ovarian cancer cell lines. Furthermore, an increase in CDK9 was significantly associated with poor prognosis in our CRC patients[10].
It was previously observed that there are differences between left and right colon cancers in molecular biological characteristics, morbidity, and response to targeted drug therapy[42]. The disease-free survival rate of resected right colon cancer and left colon cancer after radical surgery is similar, but patients with metastatic left colon cancer have a longer survival rate than patients with right colon cancer after palliative chemotherapy[2]. Some data indicate that the survival rate of right colon cancer patients has not improved since 1980 compared to left colon cancer patients, and the prognosis of these right colon cancer patients is worse[43].
It has been shown that autophagy plays a dual role in cancer: Autophagy activation can promote the survival of cancer cells (protective autophagy) or autophagy can lead to the death of cancer cells (cytotoxic/nonprotective autophagy)[43]. The TEM results showed that the level of autophagy in the right colon cancer tissue was significantly higher than that in the left colon cancer tissue, which may be one of the reasons for the poor prognosis of right colon cancer compared with left colon cancer. The specific mechanism of the differences between left and right colon cancer needs further study.
We found that the incidence of autophagy was different between left and right colon cancers with primary liver metastasis, and autophagy might be involved in the development of chemotherapy resistance. This study has laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of patients. The results from this study also show that tumor cell sensitivity to chemotherapy can be enhanced.
This work was supported by Cancer Biobank of Tianjin Medical University Cancer Institute and Hospital.
Cyclin-dependent kinase 9 (CDK9) expression and autophagy in colorectal cancer (CRC) tissues has not been widely studied. CDK9, a key regulator of transcription, may influence the occurrence and progression of CRC. The expression of autophagy-related genes BECN1 and drug resistance factor ABCG2 may also play a role in CRC.
Under normal physiological conditions, autophagy can inhibit tumorigenesis, but once a tumor forms, autophagy may promote tumor growth. Therefore, understanding the relationship between autophagy and cancer, particularly how autophagy promotes tumor growth after its formation, is a key motivation for this research.
To investigate the relationship between CDK9 expression and autophagy in CRC, assess differences in autophagy between left and right colon cancers, and analyze the associations of autophagy-related genes with clinical features and prognosis.
The research utilized a multi-faceted approach involving data analysis from The Cancer Genome Atlas database to study CDK9 expression in CRC. We also collected fresh tumor and paracarcinoma tissues for electron microscopy, immunohistochemistry, and flow cytometry to investigate autophagy. Additionally, the study analyzed the expression of autophagy-related genes BECN1 and ABCG2 and their correlation with clinical features and prognosis.
CDK9 expression was increased in CRC tissues. CDK9 expression positively correlated with the level of autophagy in colon cancer, indicating a potential link between CDK9 and autophagy in cancer progression. The incidence of autophagy was also found to be significantly higher in right colon cancer compared to left colon cancer with primary liver metastasis. Right colon cancer displayed a higher expression of ABCG2, suggesting a potential mechanism for the observed differences in prognosis between left and right colon cancers. High expression of CDK9 was found to be associated with a poor prognosis in CRC, affecting overall survival, disease-specific survival, and progression-free interval. The expression of CDK9 was linked to specific clinical features such as tumor T-stage and lymph node invasion. Finally, high expressions of ABCG2 and BECN1 were also associated with specific clinical features, and expression levels of ABCG2 had a high predictive accuracy for CRC prognosis.
CDK9 expression positively correlated with the level of autophagy in colon cancer. Thus, this study has laid the foundation for further research on the combination of CDK9 inhibitors and autophagy inhibitors in the treatment of tumor patients.
This study emphasized the significance of patient-specific treatment approaches in CRC cancer, highlighting the role of CDK9, autophagy, and drug resistance factors in prognosis. It underscored the need for combination therapies and further research to improve cancer treatment outcomes.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dambrauskas Z, Lithuania; Eid N, Malaysia S-Editor: Li L L-Editor: A P-Editor: Zhang XD
| 1. | Lee GH, Malietzis G, Askari A, Bernardo D, Al-Hassi HO, Clark SK. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol. 2015;41:300-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 267] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 2. | Shen H, Yang J, Huang Q, Jiang MJ, Tan YN, Fu JF, Zhu LZ, Fang XF, Yuan Y. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J Gastroenterol. 2015;21:6470-6478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 133] [Cited by in F6Publishing: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 3. | Tutone M, Almerico AM. Recent advances on CDK inhibitors: An insight by means of in silico methods. Eur J Med Chem. 2017;142:300-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 4. | Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5:a008904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 234] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 5. | Siemeister G, Lücking U, Wengner AM, Lienau P, Steinke W, Schatz C, Mumberg D, Ziegelbauer K. BAY 1000394, a novel cyclin-dependent kinase inhibitor, with potent antitumor activity in mono- and in combination treatment upon oral application. Mol Cancer Ther. 2012;11:2265-2273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 6. | Barnum KJ, O'Connell MJ. Cell cycle regulation by checkpoints. Methods Mol Biol. 2014;1170:29-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 300] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 7. | Martin MP, Endicott JA, Noble MEM. Structure-based discovery of cyclin-dependent protein kinase inhibitors. Essays Biochem. 2017;61:439-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Kretz AL, Schaum M, Richter J, Kitzig EF, Engler CC, Leithäuser F, Henne-Bruns D, Knippschild U, Lemke J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumour Biol. 2017;39:1010428317694304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Anshabo AT, Milne R, Wang S, Albrecht H. CDK9: A Comprehensive Review of Its Biology, and Its Role as a Potential Target for Anti-Cancer Agents. Front Oncol. 2021;11:678559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 62] [Article Influence: 20.7] [Reference Citation Analysis (1)] |
| 10. | Wang J, Dean DC, Hornicek FJ, Shi H, Duan Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in ovarian cancer. FASEB J. 2019;33:5990-6000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Lee DJ, Zeidner JF. Cyclin-dependent kinase (CDK) 9 and 4/6 inhibitors in acute myeloid leukemia (AML): a promising therapeutic approach. Expert Opin Investig Drugs. 2019;28:989-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 12. | Wu T, Qin Z, Tian Y, Wang J, Xu C, Li Z, Bian J. Recent Developments in the Biology and Medicinal Chemistry of CDK9 Inhibitors: An Update. J Med Chem. 2020;63:13228-13257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 13. | Polier G, Giaisi M, Köhler R, Müller WW, Lutz C, Buss EC, Krammer PH, Li-Weber M. Targeting CDK9 by wogonin and related natural flavones potentiates the anti-cancer efficacy of the Bcl-2 family inhibitor ABT-263. Int J Cancer. 2015;136:688-698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen H, Chen JW, Chen JK, Chen M, Chen P, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen X, Chen YH, Chen YG, Chen Y, Chen YJ, Chen YQ, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D'Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang WP, Huang YR, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang L, Jiang T, Jiang X, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SY, Lee SH, Lee SS, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li Q, Li R, Li S, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu Q, Liu RY, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu Y, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CMP, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun SY, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang D, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang HD, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang Y, Wang YJ, Wang YT, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xilouri M, Xiong Y, Xu C, Xu F, Xu H, Xu J, Xu L, Xu X, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang J, Zhang JP, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4066] [Cited by in F6Publishing: 4041] [Article Influence: 505.1] [Reference Citation Analysis (0)] |
| 15. | Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433-1437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 769] [Cited by in F6Publishing: 780] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 16. | Wilde L, Tanson K, Curry J, Martinez-Outschoorn U. Autophagy in cancer: a complex relationship. Biochem J. 2018;475:1939-1954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 17. | Chakraborty J, Caicci F, Roy M, Ziviani E. Investigating mitochondrial autophagy by routine transmission electron microscopy: Seeing is believing? Pharmacol Res. 2020;160:105097. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Hu YL, Jahangiri A, Delay M, Aghi MK. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012;72:4294-4299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 19. | Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970-2979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, Huang RY, Shen HM, Manjithaya R, Kumar AP. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 366] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 21. | Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 646] [Cited by in F6Publishing: 664] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 22. | Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol. 2013;23:620-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 365] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 23. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 43169] [Article Influence: 3320.7] [Reference Citation Analysis (4)] |
| 24. | Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927-939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2607] [Cited by in F6Publishing: 2697] [Article Influence: 141.9] [Reference Citation Analysis (0)] |
| 25. | Li A, Gao M, Liu B, Qin Y, Chen L, Liu H, Wu H, Gong G. Mitochondrial autophagy: molecular mechanisms and implications for cardiovascular disease. Cell Death Dis. 2022;13:444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 50] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 26. | Yao J, Wang J, Xu Y, Guo Q, Sun Y, Liu J, Li S, Guo Y, Wei L. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. 2022;18:1879-1897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 27. | Ding R, Jin S, Pabon K, Scotto KW. A role for ABCG2 beyond drug transport: Regulation of autophagy. Autophagy. 2016;12:737-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 28. | Shu H, Yuan B, Huang Y, Wang L, He B, Sun Q, Sun L. High expression of ABCG2 is associated with chemotherapy resistance of osteosarcoma. J Orthop Surg Res. 2021;16:85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 29. | Svirnovski AI, Shman TV, Serhiyenka TF, Savitski VP, Smolnikova VV, Fedasenka UU. ABCB1 and ABCG2 proteins, their functional activity and gene expression in concert with drug sensitivity of leukemia cells. Hematology. 2009;14:204-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Arai R, Waguri S. Improved Electron Microscopy Fixation Methods for Tracking Autophagy-Associated Membranes in Cultured Mammalian Cells. Methods Mol Biol. 2019;1880:211-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Yu S, Hu Q, Fan K, Yang C, Gao Y. CSNK2B contributes to colorectal cancer cell proliferation by activating the mTOR signaling. J Cell Commun Signal. 2021;15:383-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Zhang M, Peng Y, Yang Z, Zhang H, Xu C, Liu L, Zhao Q, Wu J, Wang H, Liu J. DAB2IP down-regulates HSP90AA1 to inhibit the malignant biological behaviors of colorectal cancer. BMC Cancer. 2022;22:561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 33. | Wang T, Zhang WS, Wang ZX, Wu ZW, Du BB, Li LY, Chen YF, Yang XF, Hao XY, Guo TK. RAPTOR promotes colorectal cancer proliferation by inducing mTORC1 and upregulating ribosome assembly factor URB1. Cancer Med. 2020;9:1529-1543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432-445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 927] [Cited by in F6Publishing: 981] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 35. | Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 36. | Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 304] [Cited by in F6Publishing: 709] [Article Influence: 177.3] [Reference Citation Analysis (0)] |
| 37. | Borden KLB. CDK9 and mTOR: trading places. Blood. 2019;133:1167-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Liao M, Wang C, Yang B, Huang D, Zheng Y, Wang S, Wang X, Zhang J, Tang C, Xu Z, He Y, Huang R, Zhang F, Wang Z, Wang N. Autophagy Blockade by Ai Du Qing Formula Promotes Chemosensitivity of Breast Cancer Stem Cells Via GRP78/β-Catenin/ABCG2 Axis. Front Pharmacol. 2021;12:659297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 39. | Wang N, Yang B, Muhetaer G, Wang S, Zheng Y, Lu J, Li M, Zhang F, Situ H, Lin Y, Wang Z. XIAOPI formula promotes breast cancer chemosensitivity via inhibiting CXCL1/HMGB1-mediated autophagy. Biomed Pharmacother. 2019;120:109519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Guo T, Liu DF, Peng SH. CDK9 is up-regulated and associated with prognosis in patients with papillary thyroid carcinoma. Medicine (Baltimore). 2022;101:e28309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Ma H, Seebacher NA, Hornicek FJ, Duan Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. EBioMedicine. 2019;39:182-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 42. | Zhong M, Xiong Y, Ye Z, Zhao J, Zhong L, Liu Y, Zhu Y, Tian L, Qiu X, Hong X. Microbial Community Profiling Distinguishes Left-Sided and Right-Sided Colon Cancer. Front Cell Infect Microbiol. 2020;10:498502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Gervaz P, Usel M, Rapiti E, Chappuis P, Neyroud-Kaspar I, Bouchardy C. Right colon cancer: Left behind. Eur J Surg Oncol. 2016;42:1343-1349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |