Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Mar 15, 2024; 16(3): 945-967
Published online Mar 15, 2024. doi: 10.4251/wjgo.v16.i3.945
Published online Mar 15, 2024. doi: 10.4251/wjgo.v16.i3.945
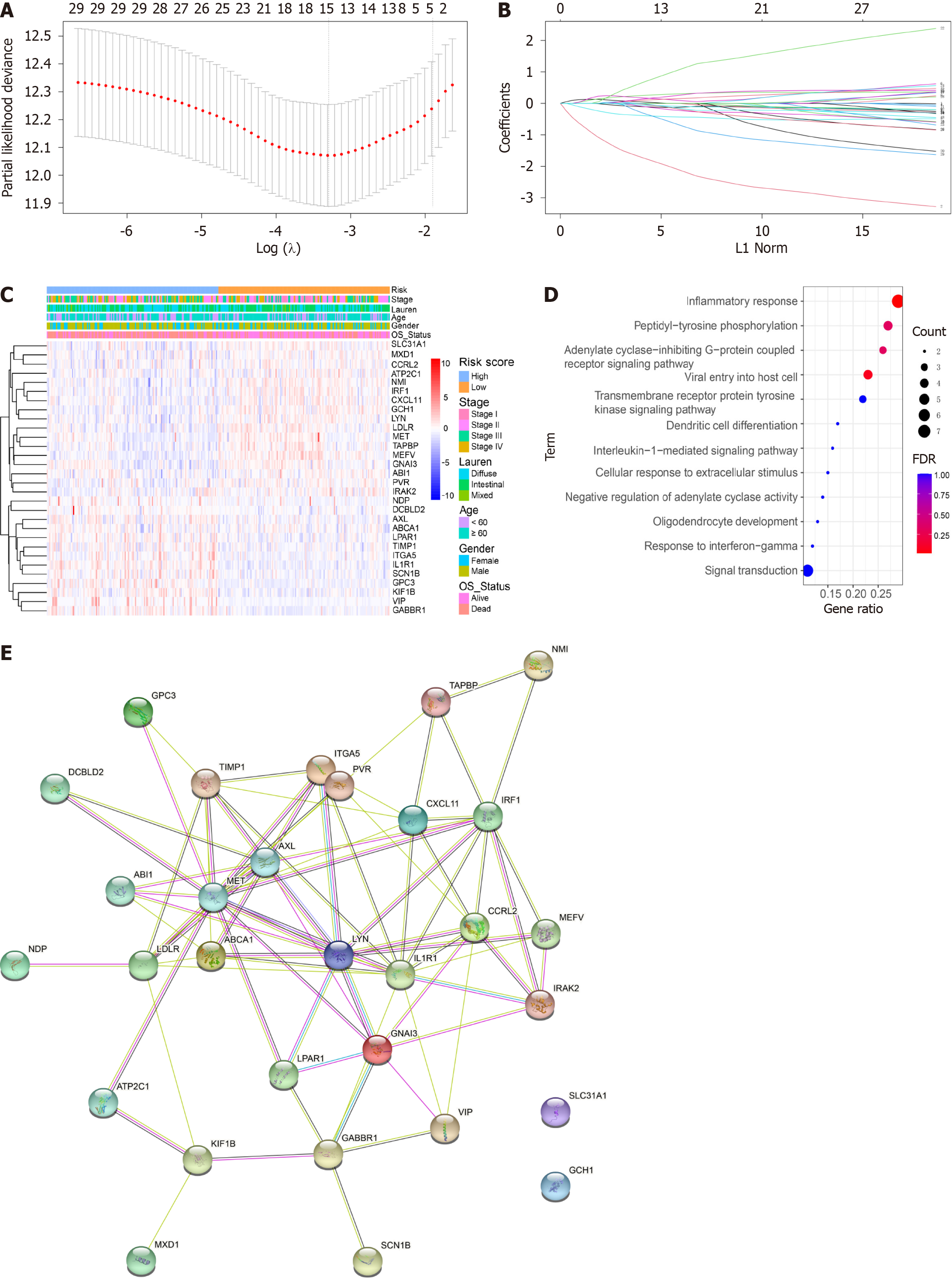
Figure 1 Identification of genes screened via least absolute shrinkage and selection operator Cox regression analysis and multivariate Cox regression analysis.
A: Partial likelihood deviance of cross-validation with different λ values for disease free survival and overall survival (OS); B: Coefficients of different genes screened via least absolute shrinkage and selection operator Cox regression analysis with different λ values for OS; C: Heatmap of inflammation-related genes and clinical features; D: Bar graph of enriched terms across enriched genes, coloured according to P value; E: PPI network of the prognostic inflammatory-related genes according to the STRING database.
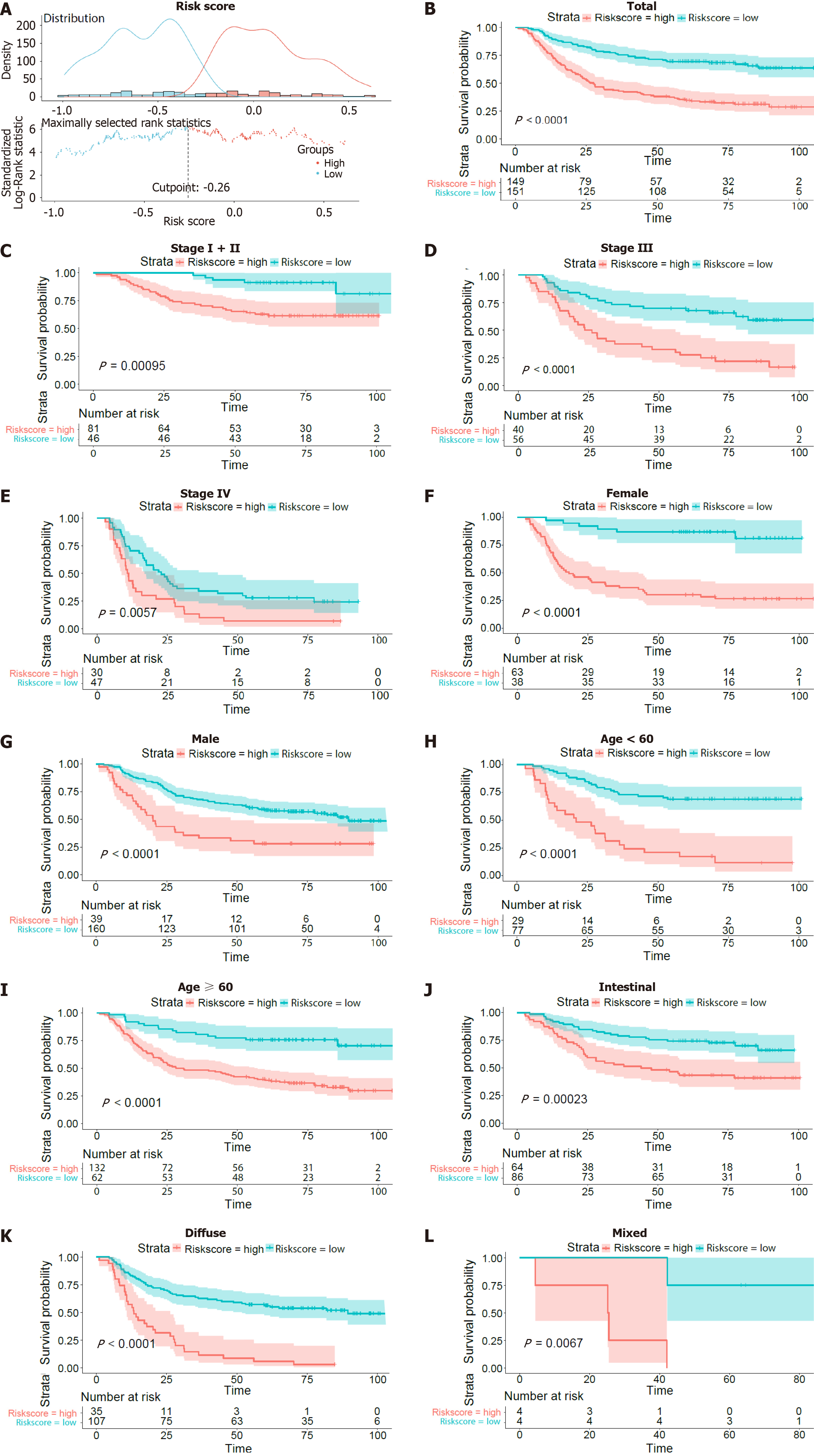
Figure 2 Survival curves of overall survival in the training dataset of GSE66229.
A: The optimal risk score cut-off point in patients with overall survival (OS) status and time records; B: K-M curves of OS between the low-risk and high-risk groups based on the total OS; C-L: K-M curves of OS stratified by TNM stage (stage I + II, stage III or stage IV), sex (female or male), age (> 60 years or ≤ 60 years), and Lauren type (intestinal or diffuse or mixed).
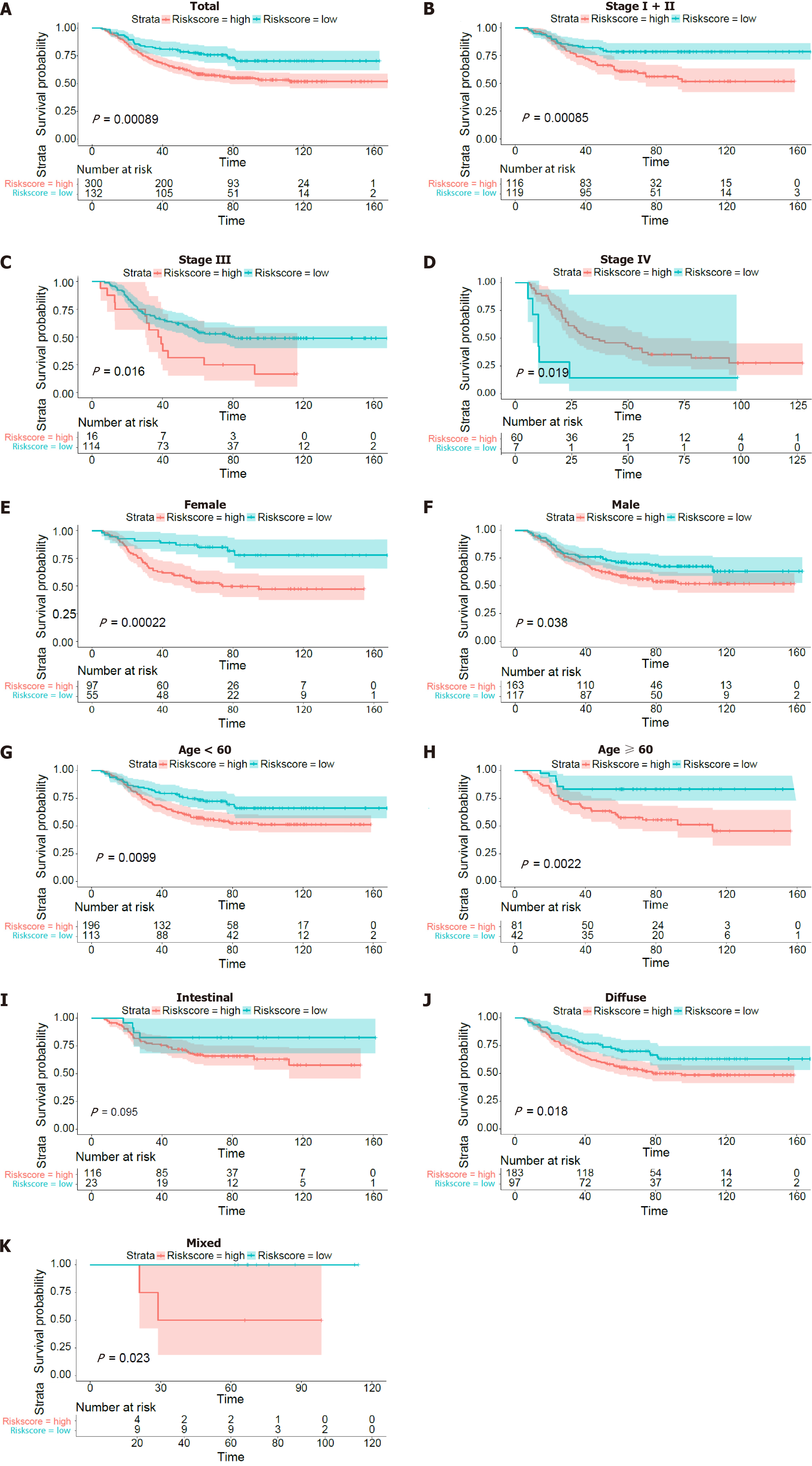
Figure 3 Kaplan-Meier curves of overall survival in patients in the GSE26253 validation dataset.
A: Kaplan-Meier curves of the overall surviving patients; B-K: Kaplan-Meier curves of overall survival stratified by TNM stage (stage I + II, stage III or stage IV), sex (female or male), age (> 60 years or ≤ 60 years), and Lauren type (intestinal or diffuse or mixed).
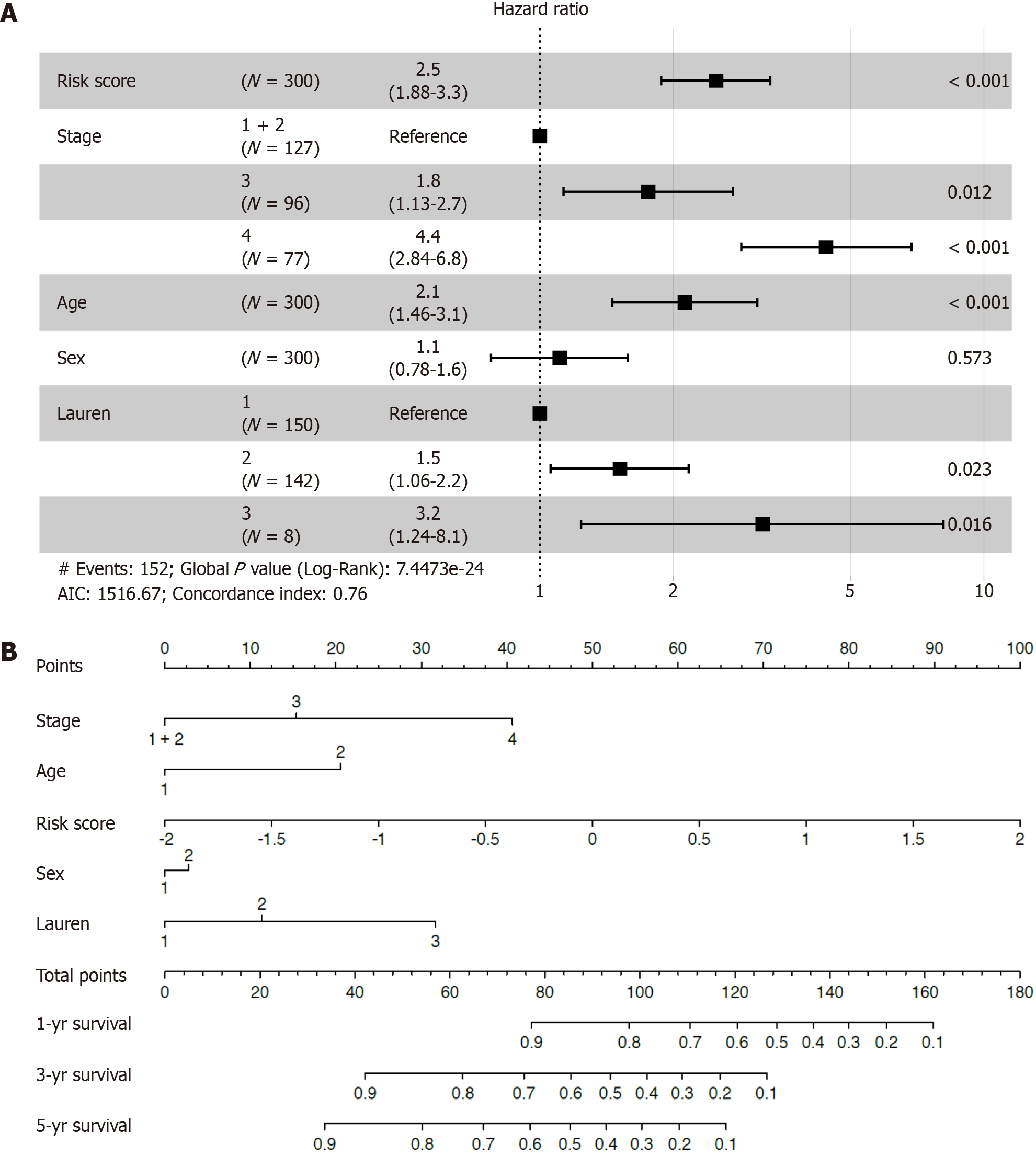
Figure 4 The risk score was an independent prognostic factor for overall survival.
A: Forest plot for overall survival (OS) in the training cohort; B: Nomogram integrating the inflammatory-related signature with clinicopathological characteristics for predicting the 1-, 3-, and 5-year OS of patients in the training cohort.
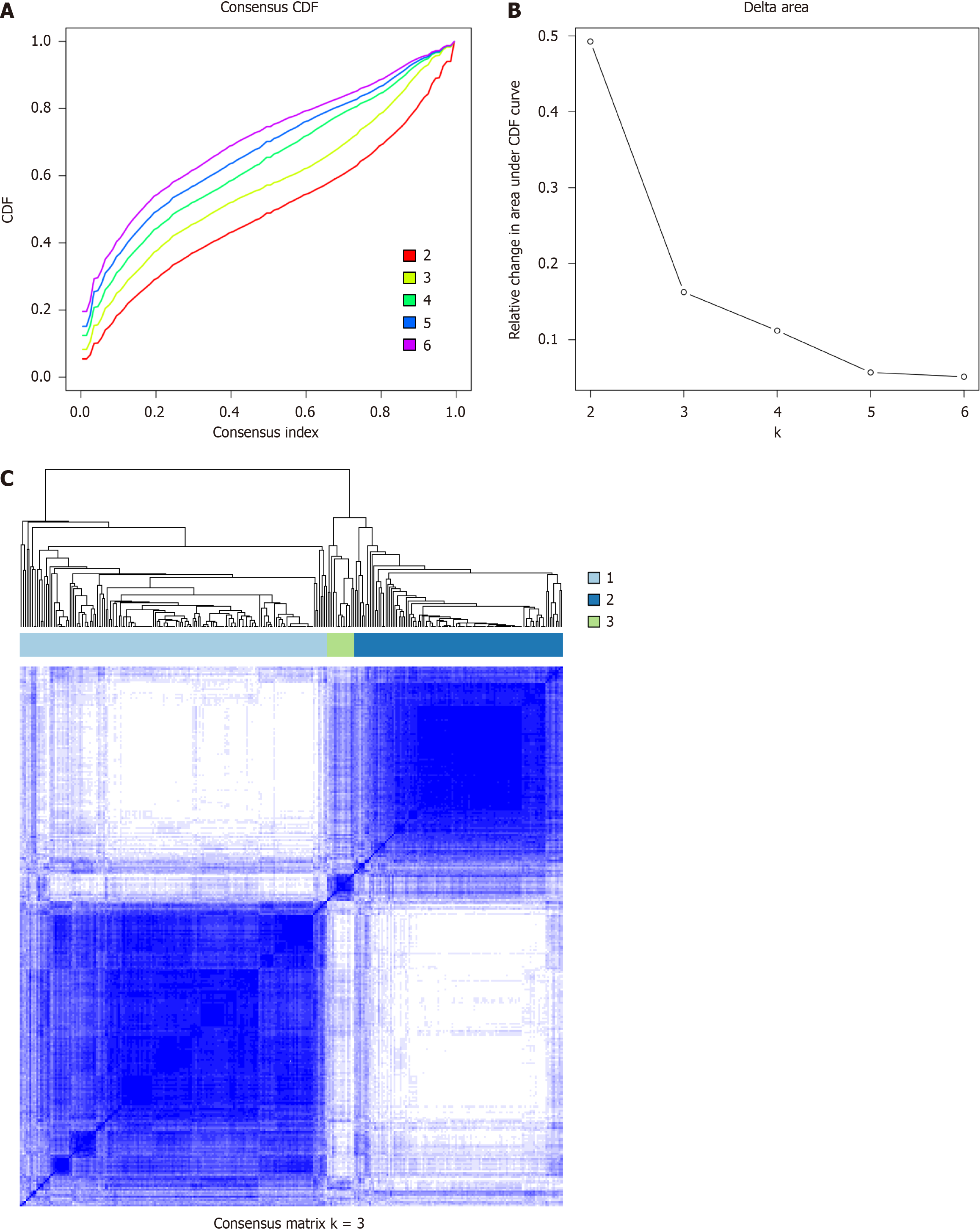
Figure 5 Identification of potential inflammatory subtypes.
A: Cumulative distribution function curve; B: Delta area of inflammatory-related genes in the training cohort; C: Heatmap depicting the consensus clustering solution (k = 3).
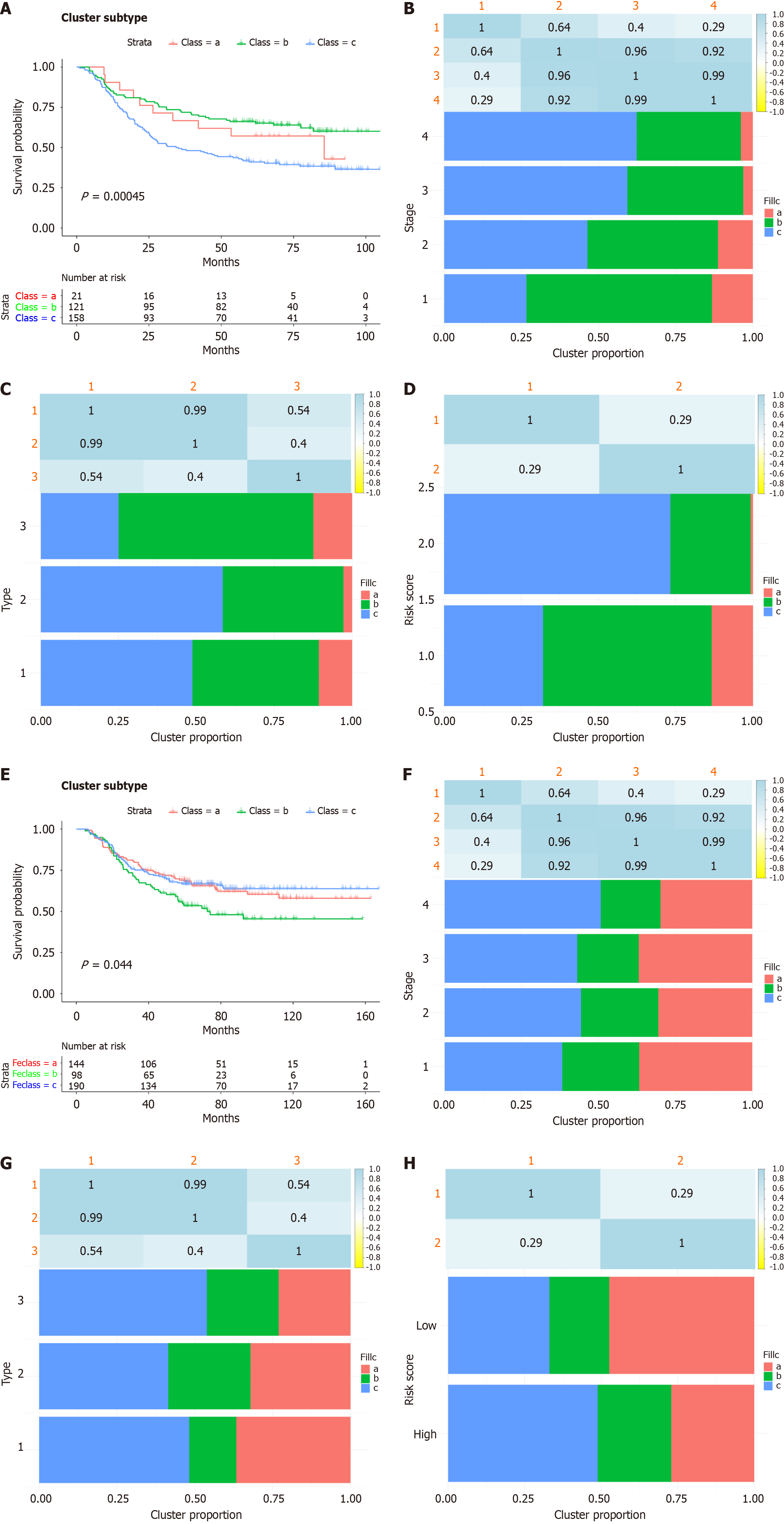
Figure 6 Subtypes of gastric cancer related to inflammatory-related genes.
A: K-M curves showing the overall survival (OS) of patients with different inflammatory subtypes in the training cohort; B-D: Distribution of FS1-FS3 expression across gastric cancer (GC) patients in the GSE66229 cohort. (B) Signature, (C) tumour stages and (D) clinically distinct subtypes in the training cohort; E: K-M curves showing the OS of patients in the validation cohort with different GC inflammatory subtypes; F-H: Distribution of FS1-FS3 across GC patients in the GSE26253 cohort, (F) signature (G) stages and (H) clinically distinct subtypes in the validation cohort.
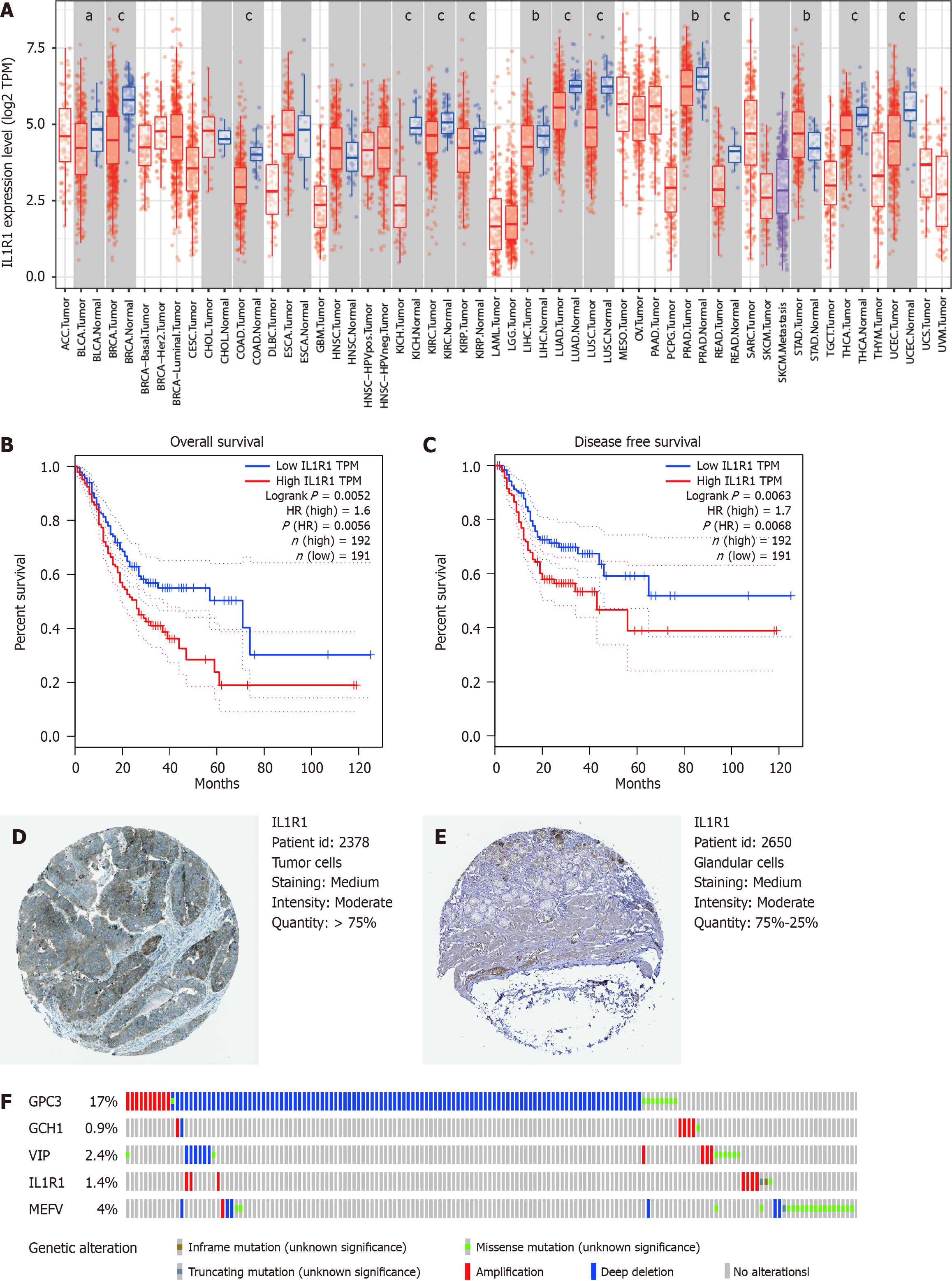
Figure 7 Expression and genetic alterations of the predictive genes.
A: The level of IL1R1 expression in different tumour types from The Cancer Genome Atlas database in Tumour Immune Estimation Resource. aP < 0.05, bP < 0.01, cP < 0.001; B: The prognostic value of IL1R1 expression for disease free survival in patients in the Gene Expression Profiling Interactive Analysis (GEPIA) cohort; C: Prognostic value of IL1R1 expression for overall survival in patients in the GEPIA cohort; D and E: Representative immunohistochemistry images of IL1R1 in gastric cancer (GC) and noncancerous gastric tissues derived from the Human Protein Atlas database; F: Genetic alterations of the five genes. The data were obtained from the cBioportal in GC patients.
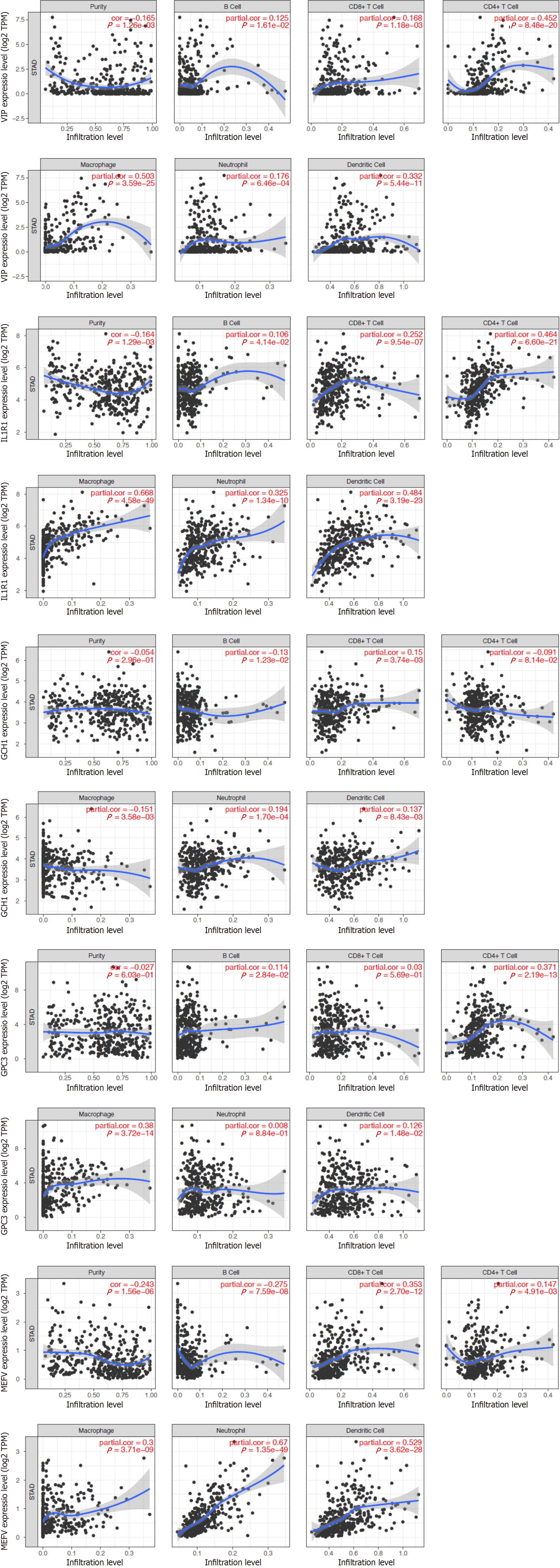
Figure 8 Relationship between the expression of inflammatory-related genes and degree of immune infiltration in gastric cancer samples, as assessed by tumour immune estimation receptor analysis.
P < 0.05 denotes statistical significance.
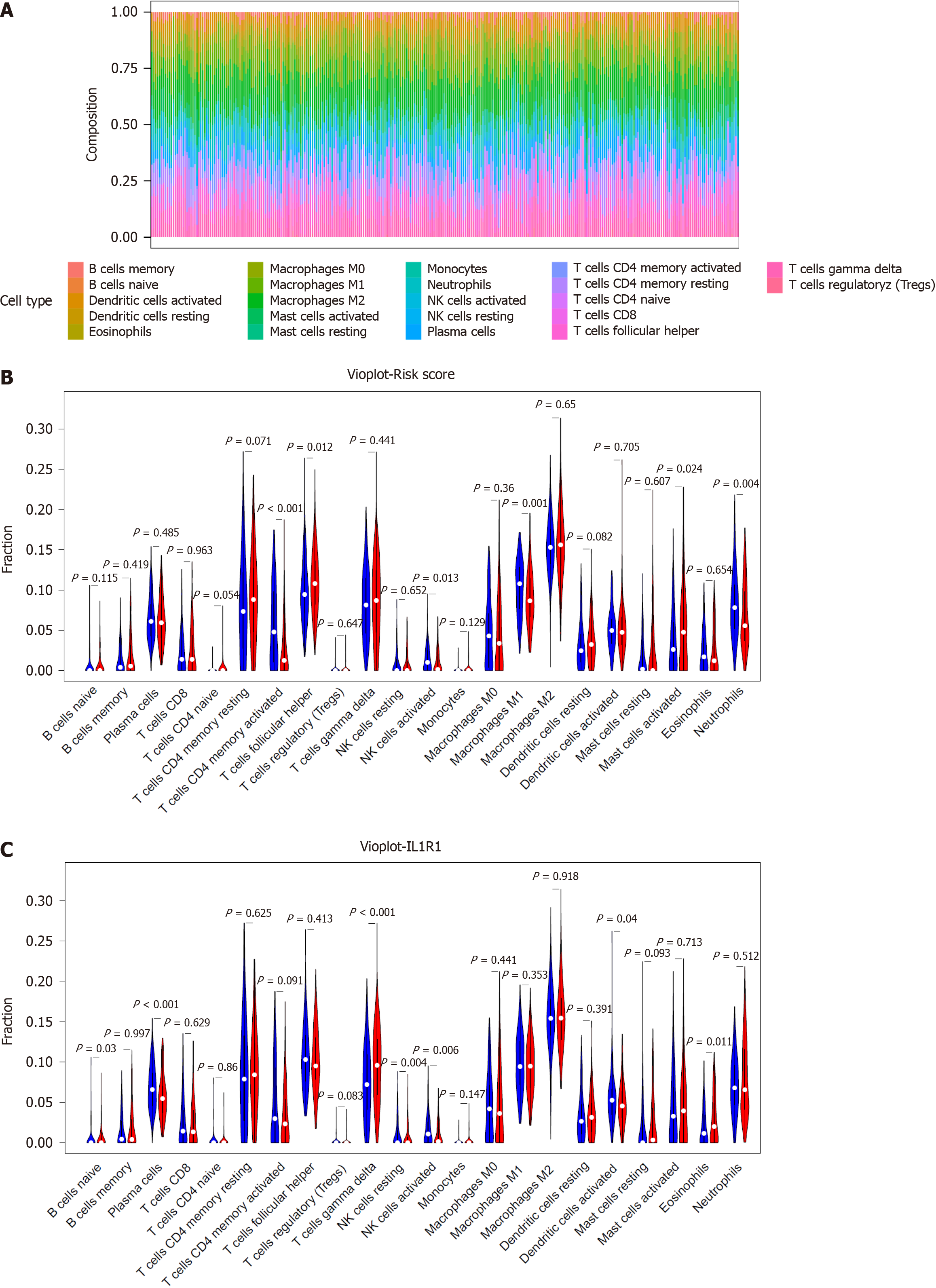
Figure 9 Correlation of the signature with immune cell infiltration.
A: Bar plots of the percentages of 22 immune cell types in the GSE66229-GC cohort. Each cell type is shown in a different colour; B: Violin plot showing the infiltration levels of 22 immune cell types in the low- and high-risk score groups. Blue: low-risk score group; red: high-risk score group; C: Violin plot showing the infiltration levels of 22 immune cell types in the low- and high-ILR1 expression groups. Blue: low-risk score expression group; red: high-risk score group.
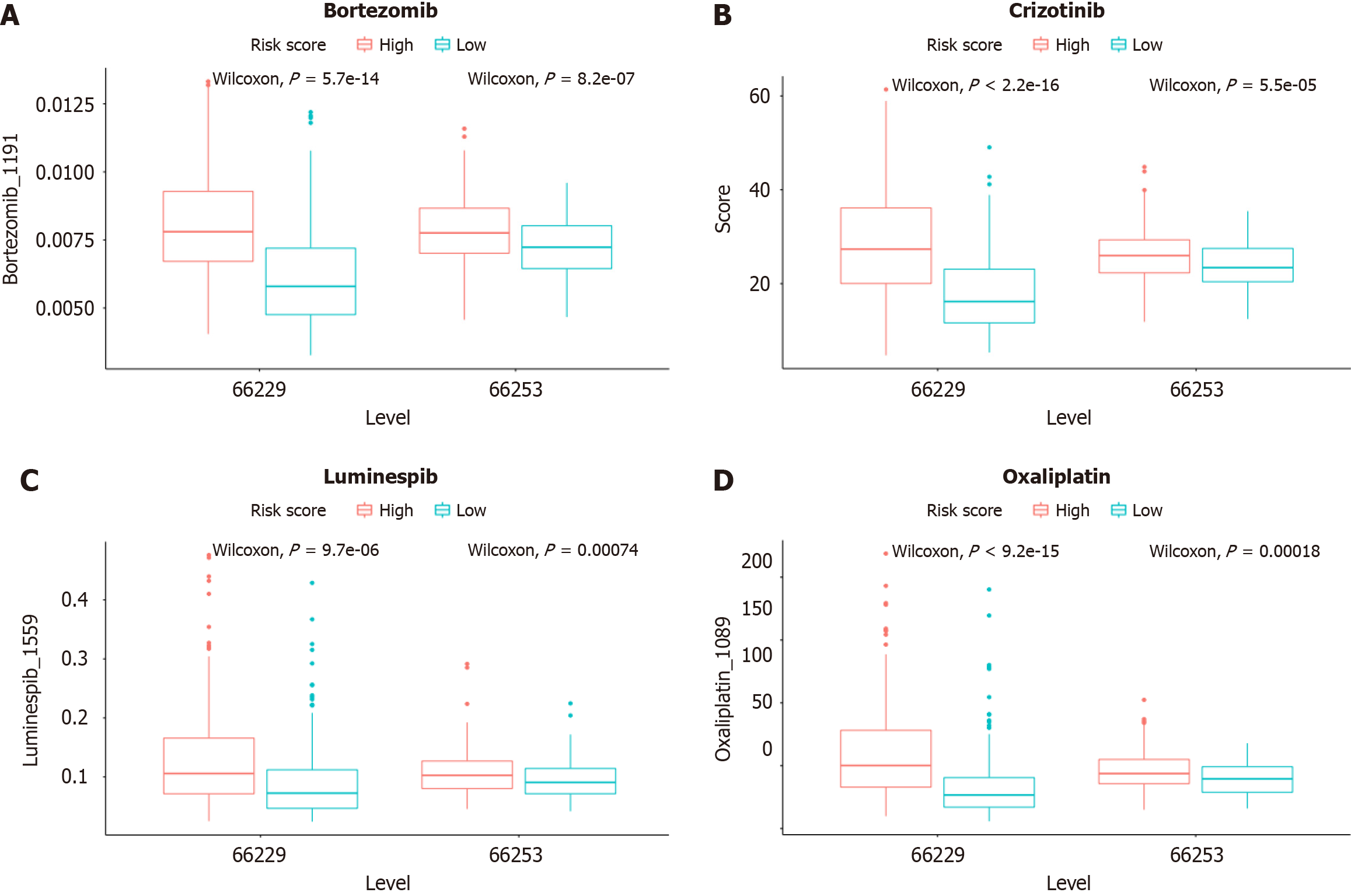
Figure 10 Prediction sensitivity of chemotherapy drugs in the high- and low-risk groups Bortezomib, Crizotinib, Luminespib, and Oxaliplatin.
- Citation: Hu JL, Huang MJ, Halina H, Qiao K, Wang ZY, Lu JJ, Yin CL, Gao F. Identification of a novel inflammatory-related gene signature to evaluate the prognosis of gastric cancer patients. World J Gastrointest Oncol 2024; 16(3): 945-967
- URL: https://www.wjgnet.com/1948-5204/full/v16/i3/945.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i3.945