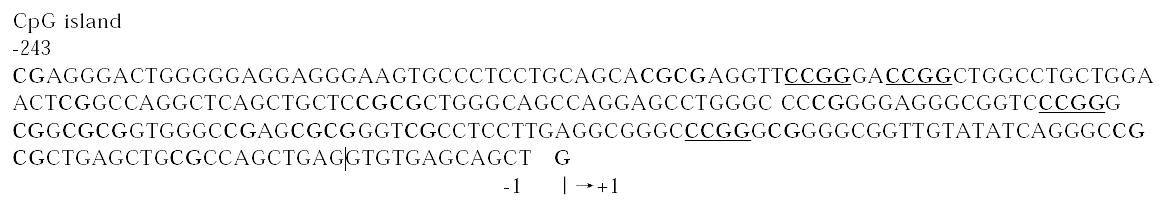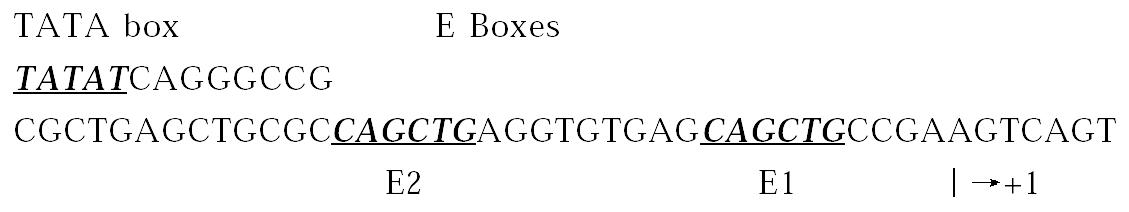Published online Jun 15, 2002. doi: 10.3748/wjg.v8.i3.400
Revised: December 22, 2001
Accepted: February 7, 2002
Published online: June 15, 2002
Cell cycle progression is regulated by interactions between cyclins and c yclin-dependent kinases (CDKs). p21WAF1 is one of the CIP/KIP family which inhib its CDKs activity. Increased expression of p21WAF1 may play an important role in the growth arrest induced in transformed cells. Although the stability of the p21WAF1 mRNA could be altered by different signals, cell differentiation and numerous influencing factors. However, recent studies suggest that two known mechanisms of epigenesis, i.e.gene inactivation by methylation in promoter region and changes to an inactive chromatin by histone deacetylation, seem to be the best candidate mechanisms for inactivation of p21WAF1. To date, almost no coding region p21WAF1 mutations have been found in tumor cells, despi te extensive screening of hundreds of various tumors. Hypermethylation of the p21WAF1 promoter region may represent an alternative mechanism by which the p21WAF1/CIP1 gene can be inactivated. The reduction of cellular DNMT prote in levels also induces a corresponding rapid increase in the cell cycle regulator p21WAF1 protein demonstrating a regulatory link between DNMT and p21WAF1 which is independent of methylation of DNA. Both histone hyperacetylation and hypoacetylation appear to be important in the carcinoma process, and induct ion of the p21WAF1 gene by histone hyperacetylation may be a mechanism by which dietary fiber prevents carcinogenesis. Here, we review the influence of hi stone acetylation and DNA methylation on p21WAF1 transcription, and affect ion of pathways or factors associated such as p53, E2A, Sp1 as well as sever al histone deacetylation inhibitors.
- Citation: Fang JY, Lu YY. Effects of histone acetylation and DNA methylation on p21WAF1 regulation. World J Gastroenterol 2002; 8(3): 400-405
- URL: https://www.wjgnet.com/1007-9327/full/v8/i3/400.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i3.400
Cell cycle progression is regulated by interactions between cyclins and CDKs[1,2]. Especially, the transition of G1 to S phase is known to be regulate d by a family of negative cell cycle regulators, CDKIs. The latter includes two families, the CIP/KIP family and the INK4 family[3-6]. p21WAF1 is one of the CIP/KIP family[7,8]. Increased expression of p21WAF1 may play a crucial role in the growth arrest induced in transformed cells[9].
p21WAF1 was first cloned and characterized as an important effector that a cted to inhibit cyclin-dependent kinase activity in p53 mediated cell cycle arrest induced by DNA damage[10,11]. It has been shown that this is a G C-rich region in the human p21WAF1 promoter[12]. Although the sta bility of the p21WAF1 mRNA could be altered by different signals cell differentiation[13] and oxidative stress[14] as well as numerous influencing factors including decorin[15], Ras/Raf protein[16], TGF-β[17] and Tax of human T cell leukemia virus type 1 (HTLV-1)[18,19]. However, two known mechanisms of epigenetic modification, gene inactivation by methylation in promoter region and changes to an inactive chromatin by hi stone deacetylation, seem to be the best candidate mechanisms for the inactivati on of CIP/KIP family[20]. In this review, we focused on the methylation, histone acetylation and some transcription factor, co-transcription factor associated with acetylation.
The post-translational modifications include acetylation, phosphorylation, meth ylation, ubiquitination and ADP-ribosylation[21]. In mammals, methylati on of the 5’ position of cytosine in the CpG dinucleotide sequence is the only n aturally occurring covalent modification of the genome. The enzyme DNA 5-cytosi ne methyltransferase (DNMT) catalyzes the transfer of a methyl group from S-adenosylmethionine to the 5 position of cytosines residing in the dinucleotide sequence CpG[22]. DNA methylation patterns correlate inversely with gene expression[23] and, therefore, DNA methylation has been suggested to be an epigenetic determinant of gene expression.
DNA methylation is believed to be an on-off switch in gene expression, CpG isla nds present in the promoter regions have been shown to be susceptible to hyperme thylation in many cancer cells[24]. CpG islands near promoters and 5’ regulatory region are usually unmethylated in normal somatic cells. In contrast, widespread methylation of CpG islands occurs in autosomal genes and leads to the silencing of the genes during oncogenic transformation.
DNA in eukaryotes is packaged with histone and non-histone proteins into chroma tin. In general, regions of chromatin that are hyperacetylated are transcription ally active, whereas regions that are hypoacetylated are silenced. Indeed, a global increase in core histone acetylation does not necessarily induce widespread transcription[25]. Histone acetylation results in charge neutralization and separation of DNA from the histones allowing nucleosomal DNA to become more accessible to transcription factors. Histone acetylation is believed to stabilize local nucleosomal structure, thereby allowing transcription factors and the ba sal trancriptional machinery access to DNA. Hyperacetylation of histones has bee n shown to mark open chromatin and to be required for trancriptional activation[26].
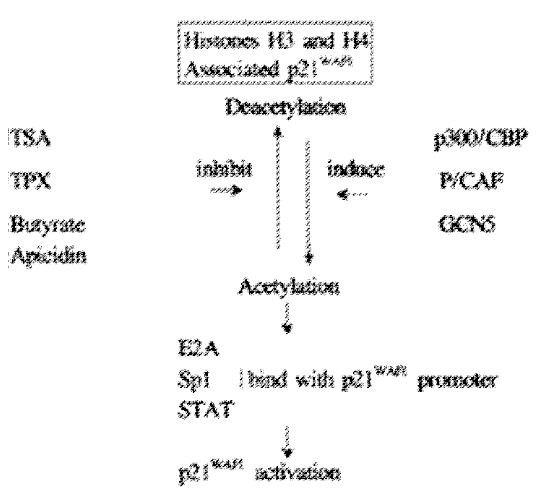
Histone acetylation is a reversible process: histone acetyltransferases (HATs) transfer the acetyl moiety from acetyl coenzyme A to the lysine neutralizes the positive charge, and histone deacetylases (HDACs) remove the acetyl groups re-establishing the positive charge in the histones. At least six human HDAC enzymes exist, and for higher eukaryotes, HDAC1 was first purified using an affinity mat rix based on the deacetylase inhibitor trapoxin[27]. HDAC inhibitor incl ude trichostatin A (TSA)[28,29], trapoxin (TPX)[30], Butyrate[31,32], MS-27-275 (a synthetic benzamide derivative)[33] and Apicidin[9,34]. Due to the inhibitory effects of the compounds of endogeno us genes that plays significant roles in G1-S progression of the cell cycle, HD AC inhibitors have been considered to be a novel class of cancer treatment agent[34].
Methylation is not genomically uniform, as unmethylated CpG are found preferenti cally in transcriptionally active chromatin. The highest density of nonmethylated CpG islands, which usually contain promoter or other regulatory DNA that is required for active transcription of a gene. CpG island chromatin is enriched in hyperacetylated histones and deficient in linker histones[35]. Recent stu dies have suggested a strong link between histone acetylation, chromatin remodeling, and gene regulation[26,36,37]. The results from many papers establi shed a link between DNA methylation, histone acetylation and sequence-specific DNA binding activity. In general, CpG island chromatin was found to contain highly acetylated histone H3 and H4. Deacetylation of histone H3 and H4 by the HDACs presumably leads to the formation of a chromatin environment that inhibits tran scription[38]. Hypoacetylated, transcriptionally silenced regions are of ten methylated[39], Furthermore, methylated DNA is transcriptionally repressed, but only under conditions in which the methylated template is assembled into uncleosomal structures[40], methylation density defines the level o f histone acetylation[41]. There are the roles of MeCP2, MBD1, MBD2, and MBD3[35], NuRD (nucleosome-remodeling histone deacetylase)[42,43] and DMAP1[44], as well as DNMT1[44,45] in the linkage of methylation with acetylation.
Usually, one could propose the negative regulation of p21WAF1 on the binding of DNMT1 with PCNA in normal cells[46], however the loss of p21WAF1 from PCNA complexes could cause abnormal gains of methylation during repair of DNA damage[47]. Moreover, the p21WAF1 gene transcription level is regulated by methylation, due to that p21WAF1 promoter contains high density of potentially methylatable CpG dinucleotides clustered around the initia tion site of transcription (Figure 1).
Dr. Nass et al[48] transfected three antisense DNMT1 (pCMV TMH) into human breast cancer MDA231 cell line, and found that the reduced DNMT1 protein and up-regulation of p21WAF1 suggesting that DNMT protein levels were inversely correlated with the level of p21WAF1 in breast cancer cells.
To date, almost no coding region p21WAF1 mutations have been found in tumor cells, despite extensive screening of hundreds of various tumors[49-51]. Hypermethylation of the p21WAF1 promoter region may represent an alter native mechanism by which the p21WAF1/CIP1 gene can be inactivated. DNMT and p21WAF1 compete for the same binding site on PCNA, an increase in DNMT expression might promote dissociation of p21WAF1 from PCNA, perhaps making p21WAF1 more susceptible to ubiquitination and proteasome degradation[52]. A decrease in DNMT expression would then be expected to have an opposite effect on p21WAF1 stability[48]. 5-Azacytidine (5-Aza-C, a demethylating agent) mediated Sp1 expression also up-regulated activities p21WAF1[53].
Rat-1 is a cell line containing wild-type p53[54]. Allan and co-w orkers found which p21WAF1 5’UTR contains a putative CpG island which is m ethylated in Rat-1 cells that used frequently to assess transformation and for apoptosis studies, the lack of p21WAF1 expression appears to be the result of hypermethylation of the p21WAF1 promoter region, as p21WAF1 protein expression could be induced by growth of Rat-1 cells in the presence of 5-aza-2-deoxycytidine (5-Aza-dC). Furthermore, sequencing analysis of bisulfi te-treated DNA demonstrated extensive methylation of cytosine residues in CpG d inucleotides in a CpG-rich island in the promoter region of the p21WAF1 gene[55]. A report showed that altered DNA methylation was present in RMS tumors and that the DNA methyltransferase expression is increased in both embryonal and alveolar subtypes of this cancer[56,57]. They think that hypermethylation of the p21WAF1 gene at the proximal STAT-binding site, xorrelates with decreased p21WAF1 expression. The p21WAF1 gene is su bjected to methylation regulation at the transcription level and is a target of aberrant methylation in RMS cells.
However, several studies indicated that the hypermethylation of p21WAF1 was not the main machineries of p21WAF1 expression regulation. Although Young et al[58] reported that cells arrested and p21WAF1 expressed by DNMT inhibition in normal human fibroblasts. Milutinovic demonstrated that i nhibition of DNMT resulted in the rapid induction of the known tumor suppressor and cell cycle regulator p21WAF1 by a mechanism that did not involve DNA methylation of the p21WAF1 promoter, in human non-small cell lung cancer c ell line, A549 cells[59]. The reduction of cellular DNMT protein levels also induced a corresponding rapid increase in the cell cycle regulator p21WAF1 protein demonstrating a regulatory link between DNMT and p21WAF1 which was independent of methylation of DNA[60]. Shin’s result showed that the promoter of the p21WAF1 gene was not been methylated in gastric cancer cells. This confirmed that methylation was not the mechanism for inactivation of p21WAF1 in gastric cancer cells[20]. In adenomatoid polyps, although DNMT1 expression coincided with the expression of other cell proliferation markers, many DNMT1-expressing cells also expressed p21WAF1. The fidelity of DNMT1 expression was further undetermined in colorectal carcinomas, in which a striking heterogeneity in DNMT1 expression, with some carcinoma cells containing very high DNMT1 levels and others containing very low DNMT1. These results indicate that human colorectal carcinogenesis is accompanied by a progressive dys regulation of DNMT1 expression and suggest that abnormalities in DNMT1 expression may contribute to the abnormal CpG dinucleotide methylation which changes the characteristic of human colorectal carcinoma cell DNA[61].
Histone deacetylation is a general mechanism for inactivation of the p21WAF1 in gastric cancer cell lines[20]. Both histone hyperacetylation and hypoacetylation appear to be important in the carcinoma process, and induction of the p21WAF1 gene by histone hyperacetylation may be a mechanism by which dietary fiber prevents carcinogenesis[31].
Regarding the correlation of histone acetylation and p21WAF1 gene expression, that HDAC inhibitor TSA, trapoxin, butyrate and apicidin induce p21WAF1 transcriptional activity involved in most studies.
TSA is originally reported to be a fungistatic antibiotic, and it appears to be a promising tool for analyzing the many functions of histone hyperacetylation in cell proliferation and differentiation. TSA can stimulate p21WAF1 express ion in HT29 cells[32].
TPX is the microbially derived cyclotetrapeptide[62], Sambucetti found t hat it increased the level of chromatin acetylation associated with histone H3 in the trapoxin-responsive region of the p21WAF1 promoter, and it activated p21WAF1 transcription that led to elevated p21WAF1 protein levels in three kinds of human tumor cells. Since the domain of the promoter that is ne cessary for TPX-mediated activation does not contain p53 binding sites, hence p21WAF1 expression upregulation by TPX is independent of p53[30].
Sodium butyrate is a short chain fatty acid produced in the human colon by bacte rial fermentation of carbohydrates[32], causes hyperacetylation of histo ne through the inhibition of HDAC. Three years ago, Archer and his coworkers showed firstly the critical importance of p21WAF1 in butyrate-mediated growth arrest was able to cause growth arrest in the human colon cancer cell line HT-29[31]. Siavoshian[32] suggested that butyrate and TSA stimulated, the p21WAF1 expression both at the mRNA and protein levels, whereas t hey induced histone H4 hyperacetylation. Butyate sensitivity requires Sp1-3 site in conjunction with the Sp1-5 site and Sp1-6[29]. Shin et al[20] indicated that the overexpression of p21WAF1 gene occurred in human gastric cancer cell lines after butyrate treatment. Butyrate increased histone H4-acetylation in human melanoma cell lines A375 and S91 and up-regulated p21WAF1 gene transcription level[63].
Apicidin is a fungal metabolite shown to exhibit antiparasitic activity by inhibition of HDAC. Han et al[64] indicated that inhibition of HDAC activity by apicidin was closely associated with monorphological change and induction of p21WAF1, although The protein levels of cyclin D1, CDK2, HDAC1 and p 53 were not affected by the addition of apicidin for 24 hrs, whereas the induction of p21WAF1 by apicidin was reversible.
Suberoylanilide hydroxamic acid (SAHA) is a hydroxamic acid-based hybrid polar compound, and it is an inhibitor of HDAC[65,66]. SAHA causes an accumulation of acrtylated histones H3 and H4 in total cellular chromatin by 2 h, which is maintained throughout 24 h of culture with increased p21WAF1 expression, but no change in chromatin associated with the actin and p27 genes, and SAHA also induces up to a 9-fold increase in p21WAF1 mRNA and protein in T24 bladder carcinoma cells. p21WAF1 by SAHA is regulated, at least in par t, by the degree of acetylation of the gene-associated histones and that this induced increase in acetylation is gene selective[66]. These studies also suggest that p21WAF1 is HDAC inhibitor and that the p21WAF1 promoter is a useful model for study in hsitone acetylation regulated transcription.
In addition, MS-27-275 inhibits HDAC and causes hyperacetylation of histones, as well as induces the expression of p21WAF1 various tumor cell lines[33].
The data above indicated that the induction of histone hyperacetylation by HDAC inhibitor is responsible for the antiproliferative activity through the crucial role of p21WAF1 in the regulation of cell cycle.
Several genes or transcriptional regulatory proteins including p300/CBP associate to p21WAF1 gene regulation.
The p21WAF1 expression may be dependent[11,67] or independent of p53 regulation[68-70]. Also, the mechanisms of p21WAF1 transcription regulation fall into two general categories: dependent or independent of the p53 gene[31]. The p21WAF1 promoter contains five natural p53 binding sites, at positions 4001, 3764, 2311, 2276, and 1391, respectively (GenBank accession number U24170)[19].
p53 gene regulates the expression of p21WAF1, and HDAC1, 2, and 3 are al l capable of downregulating p53 function, i.e., interactions of p53 and HDAC2 likely result in p53 deacetylation, thereby reducing its transcription al activity[71]. Clark and co-workers found that loss of the G1/S che ckpoint in HIV-1-infected cells may in part be due to Tat’s ability to bind p53 and sequester its transactivation activity, as seen in both in vivo an d in vitro transcription assays[72].
p21WAF1 overexpression has been seen to inhibit two critical checkpoints i n the cell cycle, G1 and G2, through both p53-dependent and -independent[74].
Up to now, four families of nuclear proteins including p300/CBP and p300/CBP-as sociated cofactors contain an intrinsic HAT activity have been confirmed that po ssess HAT activity[74-78]. Accumulating evidences suggest that p300 and CBP are adaptors for various DNA-binding transcription factors[79]. Alt hough the precise mechanism by which p300/CBP stimulates transcription remains unclear, the discovery that p300/CBP and an associated factor P/CAF have histone acetylase activities suggests that these cofactors may regulate transcriptionth rough acetylation[80]. These activities have been proposed to modify the amino-terminal tails of the core histone ptoteins in a manner that may allow for some as yet uncharacterized modification of nucleosome structure.
p300 has been found to be required for induction of p21WAF1 expression in keratinocyte differentiation[70]. Xiao and coworkers indicated the evide nces that p300 is required for TSA-induced, Sp1-mediated p21WAF1 transcription: cotransfection of p300 elevated p21WAF1 promoter activity, and thi s elevation was dependent on TSA-responsive GC-box; TSA-induced promoter activation was blocked by the introduction of p300 dominant-negative mutant into cells; Sp1- or Sp3- mediated activation was also suppressed by this p300 dominant-negative mutant[28]. Owen et al[81] demonstrated the progesterone regulated transcription of the p21WAF1 gene through Sp1 and CBP/p300. A report[82] showed that p21WAF1 stimulated trans-activatio n by p300/CBP, p21WAF1 induction of p300 results from the activity of a discrete domain in the amino-terminal half of the protein which functioned to repress transcription. they proposed a model in which p300/CBP activity might switc hed between promoters following p21WAF1 induced cell cycle arrest.
Two human homologs of GCN5 have been cloned and shown to have HAT activity[83,84]. One homolog is human p300/CBP associated factor (hP/CAF), which is a transcriptional co-activator with intrinsic histone acetylase activity, which c ontributes to transcriptional activation by modifying chromatin and transcriptio nal factors[84,95]. The second family member is hGCN5[85,86].The ability of hGCN5 to acetylate nucleosomal histones is significantly reduced relative to its activity on free histones, where it predominantly modifies histone H3 at lysine 14.
The co-activator/adaptor protein GCN5 is a conserved histone acetyltransferase, which functions as the catalytic subunit in multiple yeast transcriptional regulatory complexes.
E2A gene encodes two alternatively spliced products, E12 and E47[87,88]. The p21WAF1 promoter contains eight putative E-box consensus sequences, two of which lie between the TATA box and the transcription starting site, E2 and E1 (as Figure 2). E1 binds E47 hetero- and homodimers and E2 has mush less aff inity for E47[89], and it contains a conserved basic region responsible for DNA binding and a helix-loop-helix domain for dimerization[90].
E2A plays important roles not only in promoting cellular differentiation but als o in suppressing cells grown[89]. E2A binds to p21WAF1, so the ove rexpression of p21WAF1 may be due to the effects of E2A transcriptional fa ctor[19]. Moreover, the overexpression of E2A proteins, such as E47 has been shown to induce p21WAF1 promoter activity independent of p53 binding sites[89].
Histone H4 but not histone H3 is acetylated from the endogenous p21WAF1 pr omoter in vivo, implying that CBP/p300, and not the SAGA complex is critical in complexing with E2A and upregulation of p21WAF1 in HTLV-1 infected cells[19].
The E3 box located 130 bp upstream from the TATA box also contributes to the act ivation of p21WAF1 expression, but the E4 to E8 boxes have no effect on p21WAF1 expression[89]. E2A is shown to be upregulated in HTLV-1 in fected T cells.
Sp family of proteins comprise ubiquitous and tissue-restricted transcription f actors that bind GC-rich DNA sequences and other related GT and GA motifs through their zinc-finger domains[91].The ubiquitously expressed and closely related Sp1 and Sp3 factors have been found to regulate the promoters of severa l genes, including cell-cycle regulated genes, with Sp1 defined as a potent cooperative transcriptional activator and Sp3 as weak trans-activator or a repress or[91,92]. Sp1-binding sites appear to play a critical role in the maintenance of the methylation-free CpG island[93].Both Sp1 and Sp3 bind th e promoter of p21WAF1 gene[94].
The proximal p21WAF1 promoter contains a TATA box[81] and six Sp1 binding sites, also Sp1-1, -2, -3, -4, -5 and -6 near the TATA box[29]. p21WAF1 is Sp1 dependent promoters[95]. The region between -154 and transcription starting site contains Sp1-1, -2, -3, -4, -5, -6 binding sites[20,81]. Sp1 is a sequence-specific transcription factor that recogn izes GGGGCGGGG and closely related sequences, often referred to as GC boxes. To Sp1, at last there are at least three homologous, transcription factors in the S p1 family: Sp2, Sp3 and Sp4[96]. Xiao also reported[28] TSA-ind uced promoter activation was blocked by the introduction of p300 dominant-negative mutant into cells. Their result from gel-shift assay[29] showed that physical and functional evidence which strongly indicated that both Sp1 and Sp 3 were responsible for TSA-induced transactivation for he murine p21WAF1 promoter in NIH3T3 cells. p21WAF1 gene is one of the natural target s of HDAC inhibitors. Sp1 is a sequence-specific transcription factor that recognizes GGGGCGGGG and closely related sequences, often being referred to as GC boxes. To Sp1 at last there are at least three homologous, transcription factors i n the Sp family: Sp2, Sp3 and Sp4[96].
The GC-rich region in the six consecutive Sp1 binding sites of the p21WAF1 promoter was digested either with methylation-sensitive HpaII or with me thylation-insensitive MspI. The resulting DNA was subjected to a PCR react ion. Sp1 binding sites are the common elements that exist in the promoters of both genes[20]. Using transient reporter gene assays, Pagliuca et al[94] determined that Sp1 was a stroung activator of p21WAF1 promoter, whereas Sp3 functioned as a weak transactivator.
STAT proteins recognize and bind to the palindromic sequence TTCNNGAA[95]. Such sequences have been identified in the p21WAF1 promoter region at nt-692, -2557 and -4232, and designated as sis-inducible element (SIE)-1, -2 and -3, respectively[97]. All three SIEs have been shown to bind STA T1. Chen et al[98] indicated that hypermethylation of p21 gene at the proximal Sis-inducible element (SIE)-1, a STAT-responsive element located upstream of the p21WAF1 CpG 5-Aza-dC, demethylation at SIE-1 reactivated p21WAF1 expression. STAT could up-regulate activation of cyclin-dep endent kinase inhibitor p21WAF1[98,99].
In addition to its role in cell cycle regulation, p21WAF1 is also believed to inhibit DNA replication through its ability to bind proliferating cell nucle ar antigen (PCNA), which is required for both replicative DNA synthesis and DNA repair. However, p21WAF1 has no inhibitory effect on the DNA repair function of PCNA[100,101]. Thus, p21WAF1 may play a central role in preventing the replication of mutations incurred after exposure of cells to DNA damage.
Histone acetylation is the major mechanism for regulation of the p21WAF1 gene in most cell lines (shown as Figure 3). Both histone hyperacetylation and hy poacetylation appear to be important in the carcinoma process. The influence of methylation on p21WAF1 gene expression is dependent on differentiation of cells and tissue. It is our anticipation that induction of the p21WAF1 gene by histone hyperacetylation may become a mechanism of dietary prevention of carcinogenesis.
Edited by Wu XN
| 1. | Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 851] [Cited by in F6Publishing: 862] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 2. | Marx J. How cells cycle toward cancer. Science. 1994;263:319-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 79] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Elledge SJ, Winston J, Harper JW. A question of balance: the role of cyclin-kinase inhibitors in development and tumorigenesis. Trends Cell Biol. 1996;6:388-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2510] [Cited by in F6Publishing: 2492] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 5. | Hall M, Bates S, Peters G. Evidence for different modes of action of cyclin-dependent kinase inhibitors: p15 and p16 bind to kinases, p21 and p27 bind to cyclins. Oncogene. 1995;11:1581-1588. [PubMed] [Cited in This Article: ] |
| 6. | Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673-4682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 249] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 7. | Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650-662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 675] [Cited by in F6Publishing: 690] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 8. | Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massagué J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1570] [Cited by in F6Publishing: 1600] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 9. | Kim JS, Lee S, Lee T, Lee YW, Trepel JB. Transcriptional activation of p21(WAF1/CIP1) by apicidin, a novel histone deacetylase inhibitor. Biochem Biophys Res Commun. 2001;281:866-871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Gu Y, Turck CW, Morgan DO. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366:707-710. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 526] [Cited by in F6Publishing: 587] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 11. | el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817-825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6088] [Cited by in F6Publishing: 6217] [Article Influence: 200.5] [Reference Citation Analysis (0)] |
| 12. | Prowse DM, Bolgan L, Molnár A, Dotto GP. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J Biol Chem. 1997;272:1308-1314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 110] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | Schwaller J, Koeffler HP, Niklaus G, Loetscher P, Nagel S, Fey MF, Tobler A. Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J Clin Invest. 1995;95:973-979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 101] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Esposito F, Cuccovillo F, Vanoni M, Cimino F, Anderson CW, Appella E, Russo T. Redox-mediated regulation of p21(waf1/cip1) expression involves a post-transcriptional mechanism and activation of the mitogen-activated protein kinase pathway. Eur J Biochem. 1997;245:730-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Ständer M, Naumann U, Wick W, Weller M. Transforming growth factor-beta and p-21: multiple molecular targets of decorin-mediated suppression of neoplastic growth. Cell Tissue Res. 1999;296:221-227. [PubMed] [Cited in This Article: ] |
| 16. | Wang LG, Liu XM, Kreis W, Budman DR. The effect of antimicrotubule agents on signal transduction pathways of apoptosis: a review. Cancer Chemother Pharmacol. 1999;44:355-361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 279] [Cited by in F6Publishing: 294] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 17. | Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623-28628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 360] [Cited by in F6Publishing: 372] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 18. | Parker SF, Perkins ND, Gitlin SD, Nabel GJ. A cooperative interaction of human T-cell leukemia virus type 1 Tax with the p21 cyclin-dependent kinase inhibitor activates the human immunodeficiency virus type 1 enhancer. J Virol. 1996;70:5731-5734. [PubMed] [Cited in This Article: ] |
| 19. | de La Fuente C, Santiago F, Chong SY, Deng L, Mayhood T, Fu P, Stein D, Denny T, Coffman F, Azimi N. Overexpression of p21(waf1) in human T-cell lymphotropic virus type 1-infected cells and its association with cyclin A/cdk2. J Virol. 2000;74:7270-7283. [PubMed] [Cited in This Article: ] |
| 20. | Shin JY, Kim HS, Park J, Park JB, Lee JY. Mechanism for inactivation of the KIP family cyclin-dependent kinase inhibitor genes in gastric cancer cells. Cancer Res. 2000;60:262-265. [PubMed] [Cited in This Article: ] |
| 21. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6156] [Cited by in F6Publishing: 5914] [Article Influence: 246.4] [Reference Citation Analysis (0)] |
| 22. | Adams RL, McKay EL, Craig LM, Burdon RH. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979;561:345-357. [PubMed] [Cited in This Article: ] |
| 23. | Yeivin A, Razin A. Gene methylation patterns and expression. EXS. 1993;64:523-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 54] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1307] [Cited by in F6Publishing: 1202] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 25. | Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1910] [Cited by in F6Publishing: 1947] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 26. | Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1345] [Cited by in F6Publishing: 1369] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 27. | Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1355] [Cited by in F6Publishing: 1331] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 28. | Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J Biol Chem. 2000;275:1371-1376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 154] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 29. | Xiao H, Hasegawa T, Isobe K. Both Sp1 and Sp3 are responsible for p21waf1 promoter activity induced by histone deacetylase inhibitor in NIH3T3 cells. J Cell Biochem. 1999;73:291-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 30. | Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, Xu H, Cohen D. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem. 1999;274:34940-34947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 321] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 31. | Archer SY, Hodin RA. Histone acetylation and cancer. Curr Opin Genet Dev. 1999;9:171-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 193] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 32. | Siavoshian S, Segain JP, Kornprobst M, Bonnet C, Cherbut C, Galmiche JP, Blottière HM. Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut. 2000;46:507-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 205] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 33. | Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci USA. 1999;96:4592-4597. [PubMed] [Cited in This Article: ] |
| 34. | Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508-2518. [PubMed] [Cited in This Article: ] |
| 35. | Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451-454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1349] [Cited by in F6Publishing: 1281] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 36. | Wade PA, Wolffe AP. Histone acetyltransferases in control. Curr Biol. 1997;7:R82-R84. [PubMed] [Cited in This Article: ] |
| 37. | Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 890] [Cited by in F6Publishing: 872] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 38. | Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 636] [Cited by in F6Publishing: 707] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 39. | Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 496] [Cited by in F6Publishing: 541] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 40. | Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157-165. [PubMed] [Cited in This Article: ] |
| 41. | Schübeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, Groudine M. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol Cell Biol. 2000;20:9103-9112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 127] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 42. | Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924-1935. [PubMed] [Cited in This Article: ] |
| 43. | Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 580] [Cited by in F6Publishing: 599] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 44. | Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 761] [Cited by in F6Publishing: 758] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 45. | Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 693] [Cited by in F6Publishing: 677] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 46. | Baylin SB. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997;277:1948-1949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 145] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 47. | Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996-2000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 689] [Cited by in F6Publishing: 727] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 48. | Nass SJ, Ferguson AT, El-Ashry D, Nelson WG, Davidson NE. Expression of DNA methyl-transferase (DMT) and the cell cycle in human breast cancer cells. Oncogene. 1999;18:7453-7461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 49. | Balbín M, Hannon GJ, Pendás AM, Ferrando AA, Vizoso F, Fueyo A, López-Otín C. Functional analysis of a p21WAF1,CIP1,SDI1 mutant (Arg94 --& gt; Trp) identified in a human breast carcinoma. Evidence that the mutation impairs the ability of p21 to inhibit cyclin-dependent kinases. J Biol Chem. 1996;271:15782-15786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 50. | Malkowicz SB, Tomaszewski JE, Linnenbach AJ, Cangiano TA, Maruta Y, McGarvey TW. Novel p21WAF1/CIP1 mutations in superficial and invasive transitional cell carcinomas. Oncogene. 1996;13:1831-1837. [PubMed] [Cited in This Article: ] |
| 51. | Shiohara M, el-Deiry WS, Wada M, Nakamaki T, Takeuchi S, Yang R, Chen DL, Vogelstein B, Koeffler HP. Absence of WAF1 mutations in a variety of human malignancies. Blood. 1994;84:3781-3784. [PubMed] [Cited in This Article: ] |
| 52. | Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355-363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 248] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 53. | Periyasamy S, Ammanamanchi S, Tillekeratne MP, Brattain MG. Repression of transforming growth factor-beta receptor type I promoter expression by Sp1 deficiency. Oncogene. 2000;19:4660-4667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 54. | Ling CC, Guo M, Chen CH, Deloherey T. Radiation-induced apoptosis: effects of cell age and dose fractionation. Cancer Res. 1995;55:5207-5212. [PubMed] [Cited in This Article: ] |
| 55. | Allan LA, Duhig T, Read M, Fried M. The p21(WAF1/CIP1) promoter is methylated in Rat-1 cells: stable restoration of p53-dependent p21(WAF1/CIP1) expression after transfection of a genomic clone containing the p21(WAF1/CIP1) gene. Mol Cell Biol. 2000;20:1291-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Chen B, He L, Savell VH, Jenkins JJ, Parham DM. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 2000;60:3290-3298. [PubMed] [Cited in This Article: ] |
| 57. | Chen B, Liu X, Savell VH, Dilday BR, Johnson MW, Jenkins JJ, Parham DM. Increased DNA methyltransferase expression in rhabdomyosarcomas. Int J Cancer. 1999;83:10-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 58. | Young JI, Smith JR. DNA methyltransferase inhibition in normal human fibroblasts induces a p21-dependent cell cycle withdrawal. J Biol Chem. 2001;276:19610-19616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Milutinovic S, Knox JD, Szyf M. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1/sdi1). J Biol Chem. 2000;275:6353-6359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 86] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 60. | Fournel M, Sapieha P, Beaulieu N, Besterman JM, MacLeod AR. Down-regulation of human DNA-(cytosine-5) methyltransferase induces cell cycle regulators p16(ink4A) and p21(WAF/Cip1) by distinct mechanisms. J Biol Chem. 1999;274:24250-24256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 90] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 61. | De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855-3860. [PubMed] [Cited in This Article: ] |
| 62. | Itazaki H, Nagashima K, Sugita K, Yoshida H, Kawamura Y, Yasuda Y, Matsumoto K, Ishii K, Uotani N, Nakai H. Isolation and structural elucidation of new cyclotetrapeptides, trapoxins A and B, having detransformation activities as antitumor agents. J Antibiot (Tokyo). 1990;43:1524-1532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 150] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 63. | Demary K, Wong L, Spanjaard RA. Effects of retinoic acid and sodium butyrate on gene expression, histone acetylation and inhibition of proliferation of melanoma cells. Cancer Lett. 2001;163:103-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 64. | Han JW, Ahn SH, Park SH, Wang SY, Bae GU, Seo DW, Kwon HK, Hong S, Lee HY, Lee YW. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 2000;60:6068-6074. [PubMed] [Cited in This Article: ] |
| 65. | DiGiuseppe JA, Weng LJ, Yu KH, Fu S, Kastan MB, Samid D, Gore SD. Phenylbutyrate-induced G1 arrest and apoptosis in myeloid leukemia cells: structure-function analysis. Leukemia. 1999;13:1243-1253. [PubMed] [Cited in This Article: ] |
| 66. | Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014-10019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 878] [Cited by in F6Publishing: 856] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 67. | el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, Hamilton SR. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910-2919. [PubMed] [Cited in This Article: ] |
| 68. | Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018-1021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 895] [Cited by in F6Publishing: 900] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 69. | Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024-1027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 783] [Cited by in F6Publishing: 841] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 70. | Missero C, Calautti E, Eckner R, Chin J, Tsai LH, Livingston DM, Dotto GP. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc Natl Acad Sci USA. 1995;92:5451-5455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 281] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 71. | Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436-20443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in F6Publishing: 338] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 72. | Clark E, Santiago F, Deng L, Chong S, de La Fuente C, Wang L, Fu P, Stein D, Denny T, Lanka V. Loss of G(1)/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J Virol. 2000;74:5040-5052. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935-944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 815] [Cited by in F6Publishing: 841] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 74. | Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641-643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1337] [Cited by in F6Publishing: 1364] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 75. | Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 210] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 76. | Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 983] [Cited by in F6Publishing: 974] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 77. | Yang X, Herrmann CH, Rice AP. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576-4584. [PubMed] [Cited in This Article: ] |
| 78. | Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953-959. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2118] [Cited by in F6Publishing: 2158] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 79. | Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 320] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 80. | Martínez-Balbás MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 497] [Cited by in F6Publishing: 526] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 81. | Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21(WAF1) cyclin- dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696-10701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 279] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 82. | Snowden AW, Anderson LA, Webster GA, Perkins ND. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol Cell Biol. 2000;20:2676-2686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 105] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 83. | Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593-602. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 145] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 84. | Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1147] [Cited by in F6Publishing: 1226] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 85. | Schiltz RL, Nakatani Y. The PCAF acetylase complex as a potential tumor suppressor. Biochim Biophys Acta. 2000;1470:M37-M53. [PubMed] [Cited in This Article: ] |
| 86. | Smith ER, Belote JM, Schiltz RL, Yang XJ, Moore PA, Berger SL, Nakatani Y, Allis CD. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948-2954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 87. | Mahajan MA, Park ST, Sun XH. Association of a novel GTP binding protein, DRG, with TAL oncogenic proteins. Oncogene. 1996;12:2343-2350. [PubMed] [Cited in This Article: ] |
| 88. | Peverali FA, Ramqvist T, Saffrich R, Pepperkok R, Barone MV, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291-4301. [PubMed] [Cited in This Article: ] |
| 89. | Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888-5896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 262] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 90. | Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777-783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1812] [Cited by in F6Publishing: 2034] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 91. | Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313-1323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 238] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 92. | Birnbaum MJ, van Wijnen AJ, Odgren PR, Last TJ, Suske G, Stein GS, Stein JL. Sp1 trans-activation of cell cycle regulated promoters is selectively repressed by Sp3. Biochemistry. 1995;34:16503-16508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 156] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 93. | Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 555] [Cited by in F6Publishing: 585] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 94. | Pagliuca A, Gallo P, Lania L. Differential role for Sp1/Sp3 transcription factors in the regulation of the promoter activity of multiple cyclin-dependent kinase inhibitor genes. J Cell Biochem. 2000;76:360-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 95. | Periyasamy S, Ammanamanchi S, Tillekeratne MP, Brattain MG. Repression of transforming growth factor-beta receptor type I promoter expression by Sp1 deficiency. Oncogene. 2000;19:4660-4667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 96. | Hagen G, Müller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519-5525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 444] [Cited by in F6Publishing: 481] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 97. | Horvath CM, Wen Z, Darnell JE. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984-994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 391] [Cited by in F6Publishing: 410] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 98. | Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719-722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 608] [Cited by in F6Publishing: 620] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 99. | Darnell JE. STATs and gene regulation. Science. 1997;277:1630-1635. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2976] [Cited by in F6Publishing: 3003] [Article Influence: 111.2] [Reference Citation Analysis (0)] |
| 100. | Li R, Waga S, Hannon GJ, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 463] [Cited by in F6Publishing: 502] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 101. | Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574-578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1186] [Cited by in F6Publishing: 1230] [Article Influence: 41.0] [Reference Citation Analysis (0)] |